Figure 1.
DHRSX variants in a presumptive pseudoautosomal-recessive congenital disorder of glycosylation
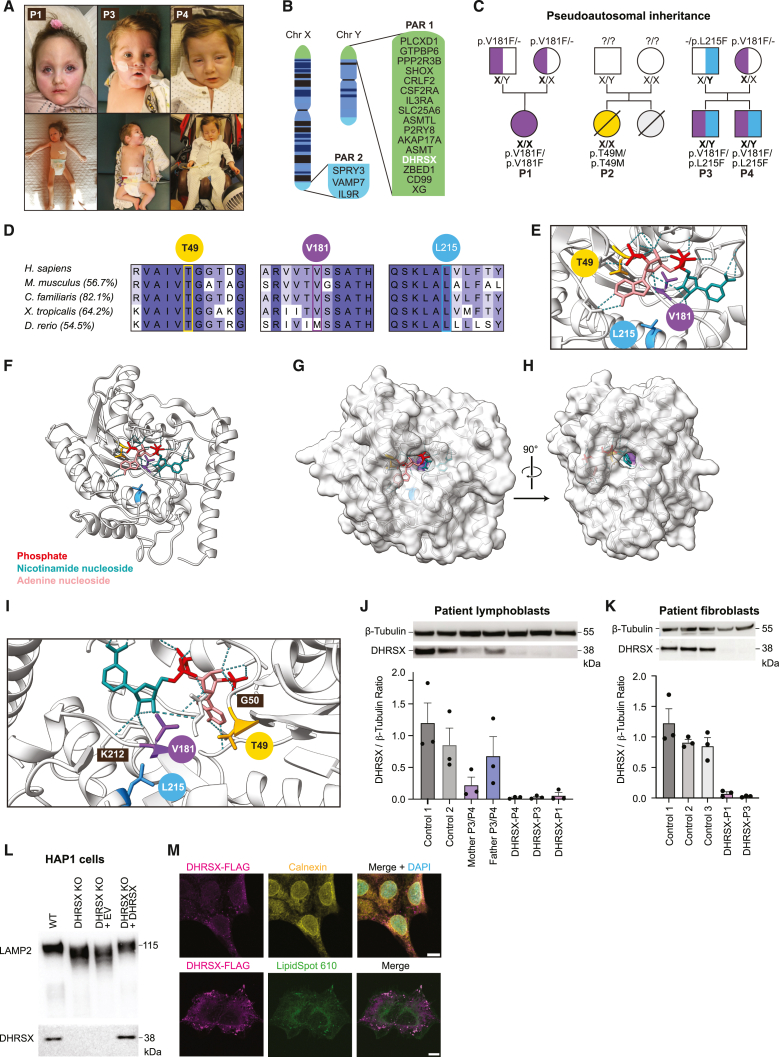
(A) Patients 1, 3, and 4 carrying DHRSX variants.
(B) The pseudoautosomal regions (PAR) of the X and Y chromosomes.
(C) Pedigree displaying the inheritance of DHRSX variants. Boldface indicates the location of DHRSX variants.
(D) Conservation of DHRSX amino acids affected by variants, and % sequence identity of the entire protein sequences as determined by ClustalW.
(E) 3D models produced by Alphafill optimization of the Alphafold Q8N5I4 model to contain NADP+ at the predicted active site. In pink the adenine nucleoside moiety, in red the phosphate groups, in turquoise the nicotinamide nucleoside moiety.
(F) Alphafold/Alphafill model showing the proximity of the amino acids substituted in patients 1–4 to the predicted active site containing NADP+. Thr49, yellow; Val181, purple; Leu215, blue.
(G) 3D surface model showing the position of the predicted active site with NADP+ bound.
(H) Rotation of the 3D surface model showing the channel presumably allowing access of lipid substrates to the active site.
(I) Localization of Thr49 (yellow), Val181 (purple), and Leu215 (blue) showing proximity to the predicted active site.
(J) Western blot analysis of DHRSX protein levels in EBV-immortalized lymphoblasts from controls, parents, and patients. Bar graphs represent DHRSX protein levels normalized to β-tubulin (mean ± SEM, n = 3).
(K) DHRSX protein levels in dermal fibroblasts from three controls, and patients with DHRSX variants (patients 1 and 3). Data are presented as in (J).
(L) Western blot analysis of LAMP2 mobility and DHRSX in wild-type (“WT”) and DHRSX KO HAP1 cells at baseline, and upon transduction with a lentiviral vector driving expression of DHRSX (“+DHRSX”) or an empty cassette (“EV”).
(M) Immunofluorescence of HAP1 cells expressing DHRSX with a C-terminal FLAG tag, using anti-FLAG for DHRSX, anti-calnexin as ER marker, lipid droplet stain LipidSpot 610, and the nuclear counterstain DAPI. Scale bars, 10 μm.
See also Figure S1.