Figure S1.
Analysis of the function of DHRSX, related to Figure 1
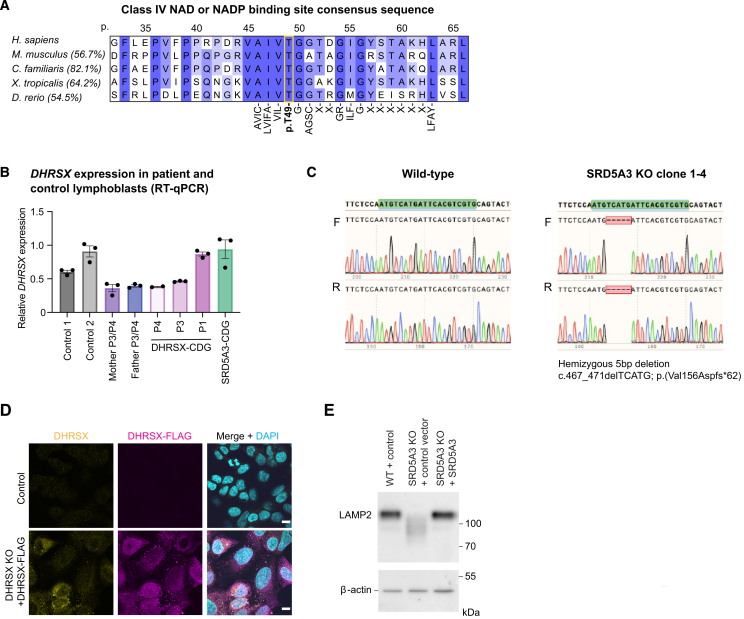
(A) Conservation of the predicted Class IV NAD or NADP binding site consensus sequence ([AVIC]-[LVIFA]-[VIL]-T-G-[AGSC]-X2-GR-ILF-G-X6-[LFAY]) in the indicated vertebrate species. Between brackets are the % sequence identity of the entire protein-coding sequences as determined by ClustalW alignment. Amino acids indicated under the consensus sequence are those required for either NAD or NADP binding as part of the class IV motif. T49 is obligatory. Amino acid positions indicated above the sequence relate to those in the human DHRSX sequence (Q8N5I4).
(B) Expression of DHRSX mRNA in EBV-immortalized lymphoblasts from controls (C1, C2), DHRSX-CDG patients (P1, P3, P4), the parents of P3/P4, and an SRD5A3-CDG patient as measured by RT-qPCR. Results are normalized to the expression of HPRT1 and then to the mean of controls. Data are represented as the mean of three biological replicates ±SEM.
(C) Sanger sequencing analysis showing a hemizygous 5bp deletion c.467_471 delTCATG; p.(Val156Aspfs∗62) in SRD5A3, confirming gene deletion (KO).
(D) Immunofluorescence analysis of WT HAP1 cells (control), and DHRSX KO HAP1 cells stably transfected with an expression construct for human DHRSX with a C-terminal triple FLAG tag. Labeling with anti-DHRSX (yellow), anti-FLAG DHRSX-FLAG (magenta) and DAPI (cyan) confirms staining in lipid droplet-like structures, and specificity of the anti-FLAG signal. Scale bars = 10 μm.
(E) Western blot analysis shows increased LAMP2 mobility indicative of hypoglycosylation in SRD5A3 KO HAP1 cells. Stable re-expression of WT SRD5A3 led to a migration of LAMP2 comparable to the one seen in WT HAP1 cells.