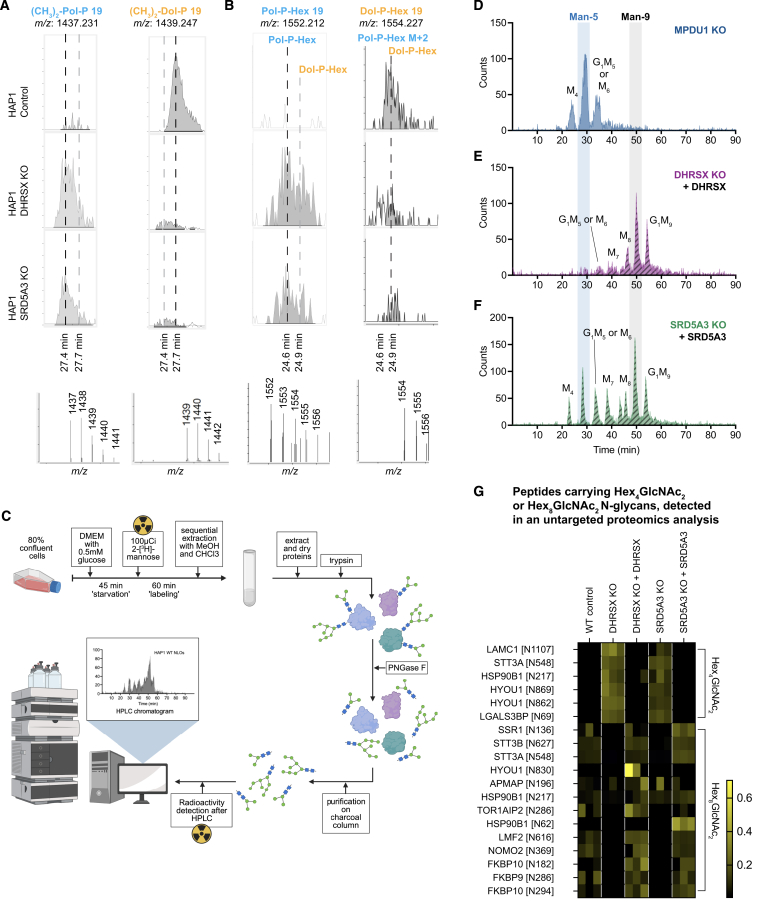
Figure S6.
Additional data corroborating effects on polyisoprenoid adducts and glycosylation, supporting Figure 6
(A) Representative mass spectra and extracted ion chromatograms for dimethylated dolichol-phosphate and polyprenol-phosphate (NH4+ adducts in positive mode) acquired in samples from control, DHRSX KO and SRD5A3 KO HAP1 cells.
(B) Representative mass spectra and extracted ion chromatograms (in negative mode) for Dolichol-phosphohexose and polyprenol-phosphohexose in control, DHRSX KO and SRD5A3 KO HAP1 cells. Dolichol-P-hexose and Polyprenol-P-Hexose represent a mixture of mannose and glucose derivatives.
(C) Experimental setup of analysis of newly synthesized N-linked oligosaccharides with radioactive mannose in HAP1 cells. Cells were grown to 90% confluency in a T25 flask, then underwent a glucose deprivation step followed by 1 h of labeling with 100 μCi tritiated 23[H]Mannose. Cells then underwent sequential extraction with chloroform and methanol, then glycoproteins were purified from the resulting protein pellet. After overnight digestion steps with trypsin and PNGase F, N-linked oligosaccharide (NLO) extracts were injected and analyzed by HPLC.
(D) Newly synthesized N-linked oligosaccharides (NLO) were detected by HPLC after incubation of MPDU1 KO HAP1 cells labeled with 100 μCi 23[H]Mannose, showing characteristic accumulation of Man4, Man5 and Glc1Man5/M6 species and deficiency of Man9 species.
(E and F) Newly synthesized NLOs were detected by HPLC after incubation of DHRSX KO HAP1 cells complemented with WT DHRSX (E), or SRD5A3 KO HAP1 cells complemented with WT SRD5A3 (F) with 100 μCi tritiated 23[H]Mannose, showing a restoration of full-length Man9 species.
(G) N-linked Hex4GlcNAc2 (corresponding to Man-4) and Hex8GlcNAc2 (corresponding to Man-8) were identified and quantified by an untargeted proteomics approach in membrane extracts of WT, DHRSX KO and SRD5A3 KO HAP1 cells and their respective complementations. The abundance of the indicated peptides was normalized to the abundance of the corresponding parent proteins. To increase visibility of differences between conditions, data are presented normalized within each modified peptide.