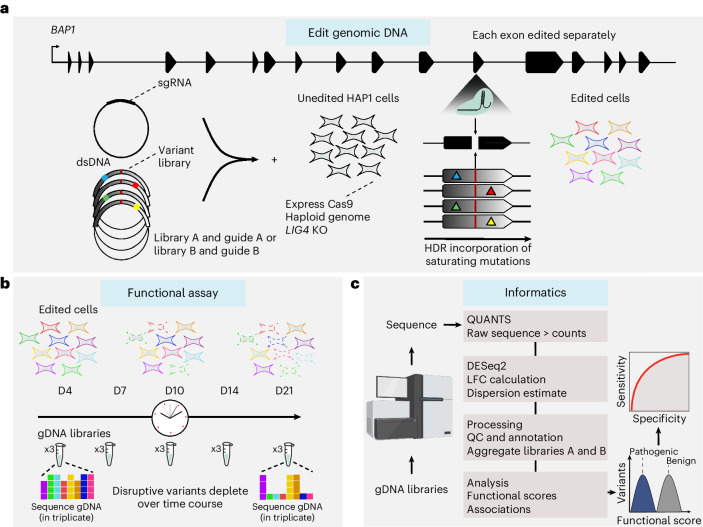
Fig. 1. Experimental design and workflow for SGE of the BAP1 locus.
a, Target regions of ≤245 bp were designed for all coding exons of the canonical BAP1 transcript: ENST00000460680.6 (ref. 52). Target regions were processed in separate experiments to sequentially cover all regions. For each region, LIG4-KO, Cas9-expressing HAP1-A5 cells were transfected in triplicate with an sgRNA-expressing plasmid and a corresponding variant library; homologous recombination with this template library at the Cas9 lesion/cut site results in the introduction of variants into the genome, generating populations of edited cells. This allows for assessment of variant function, because only benign variants will rescue cell fitness following CRISPR−Cas9-mediated disruption of BAP1, an essential gene. Each region was edited separately using two independent template library−sgRNA pairs; each variant library (library A or library B) contained saturating mutations (colored squares) and library-specific synonymous PPEs (dark red line) to prevent sgRNA−Cas9-mediated recutting of incorporated genomic tracts. dsDNA, double-stranded DNA. b, Cells were cultured over time with pellets collected at D4, D7, D10, D14 and D21. gDNA, genomic DNA. c, Sequencing was used to assess the population dynamics of genomic DNA libraries, generating counts for each variant using the QUANTS pipeline. DESeq2 was used to convert counts into an LFC of variant abundance over time. LFCs were then median scaled and a single functional score was computed through the aggregation of library A and library B data. Functional scores were categorized on the basis of a significance threshold and assessed for accuracy against variants with known pathogenic or benign classifications.