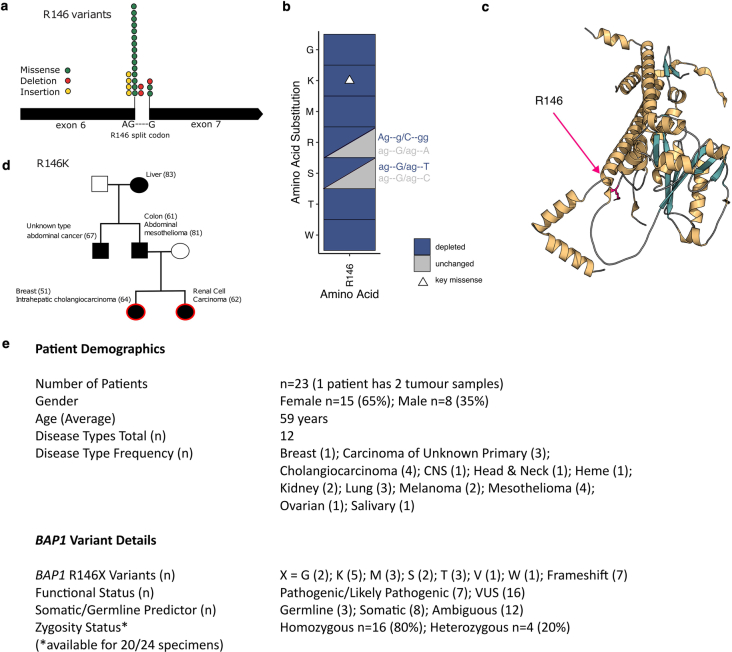
Extended Data Fig. 9. SGE resolves pathogenicity of variants in a recurrently mutated BAP1 codon identified through large-scale next generation sequencing of tumors.
Analysis of R146 variants in the Foundation Medicine cohort. We searched the Foundation Medicine database to identify novel BAP1 variants. This analysis revealed multiple variants in the R146 codon of BAP1 including missense and frameshift events. a. Variants are shown against a BAP1 gene structure and are split between different nucleotides within the codon spanning exons 6 and 7. b. All missense variants found at position R146 in the Foundation Medicine database are significantly depleted in SGE as seen by heatmap, the white triangle highlights R146K, of interest in ‘d’. Two codons were measured redundantly at the nucleotide level, and have different classifications (triplet codes in blue/grey), this includes synonymous changes, indicating disruption to splicing over the split-codon. c. All altered residues at 146 fall into a side chain proximal to the catalytic core (R146 residue highlighted in pink). d. A BAP1 R146K (c.437G>A) variant, observed in the Foundation Medicine database is a confirmed germline variant. A patient presenting with cholangiocarcinoma at 64 and their sister (a renal cell carcinoma (RCC) patient at 62) were found to carry the variant (red circles). Other first and second-degree relatives were reported to present with RCC, mesothelioma, melanoma, liver cancer, colon cancer, and a cancer of unknown primary e. Summary of patient demographics and variant details for Foundation Medicine BAP1 accessions.