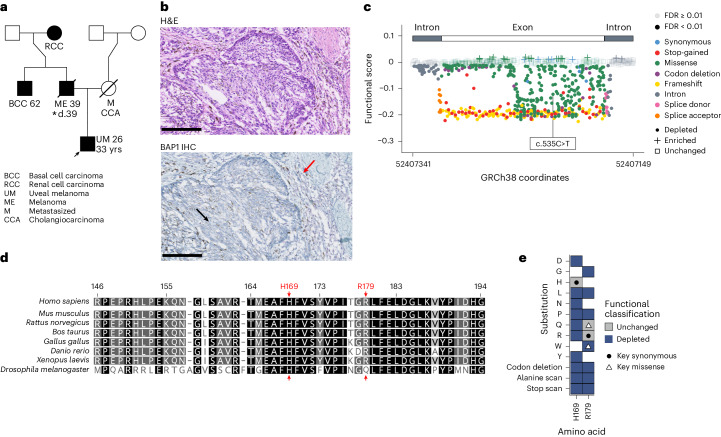
Fig. 6. Integration of the BAP1 SGE functional score with a clinical example.
a, Pedigree with a proband carrying a c.535C>T variant (HGVSc, ENST00000460680.6:c.535C>T; HGVSp, ENSP00000417132.1:p.Arg179Trp; R179W) in exon 7 of BAP1. The proband was a 33-year-old male presenting with uveal melanoma (UM) at 26 years (arrow) whose father, uncle and grandmother presented with melanoma (ME), basal cell carcinoma (BCC) and renal cell carcinoma (RCC), respectively. The proband’s mother was not known to be a carrier and died of metastatic (M) cancer, possibly cholangiocarcinoma (CCA). The pedigree follows established nomenclature: black, clinically confirmed disease (malignant tumor); square, male; circle, female; diagonal line, deceased; d., age at death; number, age at disease presentation. An asterisk indicates the patient for whom samples are shown in b. b, Pathology of the primary cutaneous melanoma in the patient from a. Top, micrograph showing hematoxylin and eosin (H&E) staining. Bottom, micrograph showing BAP1 immunohistochemical staining; staining is absent in tumor tissue (black arrow) but is present (purple cells) in immune infiltrate (red arrow). Scale bars, 100 μm. Micrographs are representative of three histological sections. c, Functional scores across exon 7. Exonic/intronic ranges within the target region are shown, with points colored by VEP consequence. Transparency based on FDR. Shape denotes functional classification. The variant in a is labeled. d, Multiple-sequence alignment of exon 7 created by global alignment of BAP1 orthologs from eight species (gap open/extension penalty = 12/3); numbers are protein positions of human BAP1 (ENSP00000417132.1) and residues are colored by identity (black, 100%; dark gray, 80−100%; light gray, 60−80%; white, <60%). R179 (and the highly conserved H169 proton donor) is highlighted by a red arrow. Note that the glutamine residue in Drosophila aligns at human position R179, the only missense variant at this position tolerated in SGE. e, Heat map (see Extended Data Fig. 6 for the full heat map) of amino acid substitutions for two key positions, H169 and R179, colored by functional classification. White space results from SNV saturation not producing all amino acid substitutions. c.535C>T produces R179W, which is depleted. R179R, a synonymous change, is unchanged, other missense changes (R179P/L/G/A/*) and R179 codon deletion are depleted and only R179Q is tolerated. H169 in the catalytic core is intolerant to all observed changes, except for a synonymous change. Black circle, key synonymous changes; white triangle, key missense changes.