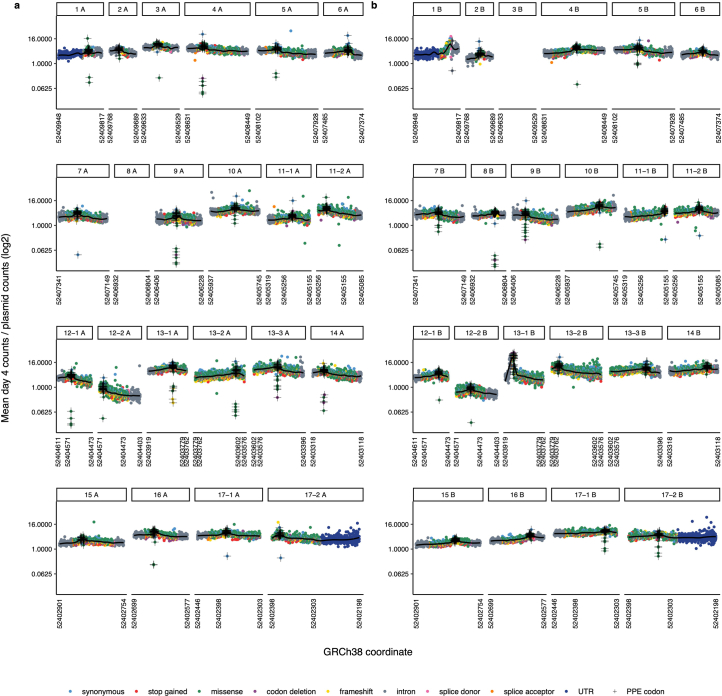
Extended Data Fig. 3. Positional effect modelling of editing across all SGE regions reveals minimal bias.
a. Counts for Day 4 Library A genomic editing events divided by plasmid library A counts for each variant as an indication of the relative rate of variant incorporation across editing regions. Variants that are edited at codons with PAM/protospacer protection edits (PPEs) are highlighted with a black cross. As expected, variants which revert the PPEs to the wild-type nucleotide sequence cannot prevent re-cutting and show depletion. Points are coloured by VEP mutational consequence. There is no extreme bias in the representation of stop-gained and frameshift variants at Day 4, indicating that compromising variants have yet to deplete by the baseline. A slight increase in incorporation is seen at most PPE positions, this is likely due to enhanced protection from re-editing relative to the rest of the edited regions. The rate of incorporation is slightly reduced at increasing distance from the Cas9 cleavage site (which is in close proximity to PPE labelled codons). Exon 8 A is missing as this guide resulted in profound cell death at the transfection stage. b. Counts for Day 4 Library B genomic editing events divided by plasmid library B counts. An extreme positional effect is observed for region 13-1 B, this data was excluded from analyses. Exon 3 B is missing as this guide resulted in profound cell death at the transfection stage.