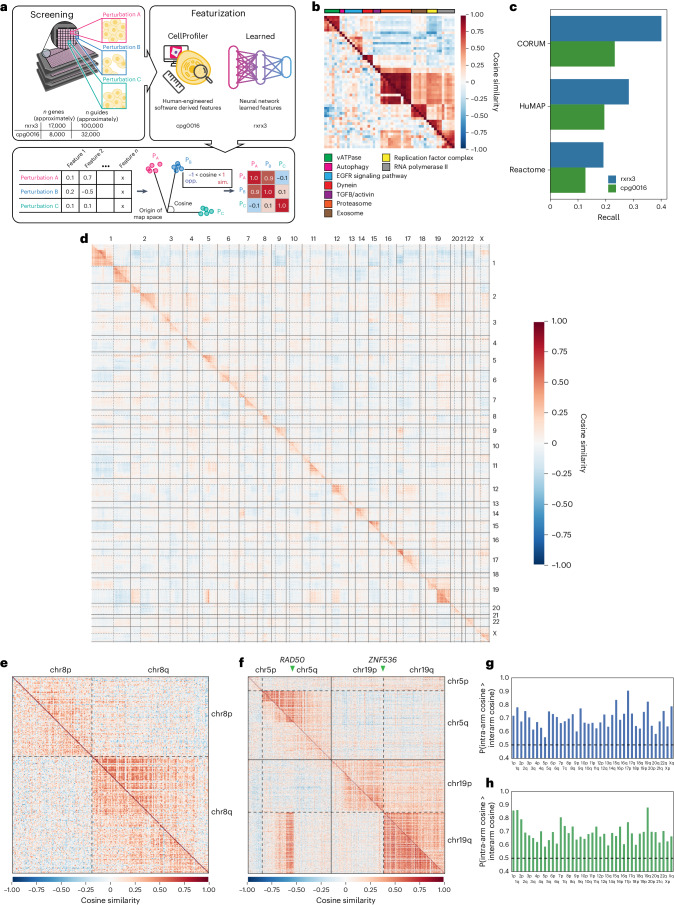
Fig. 1. Heat maps of phenotypic similarity between gene knockouts recapitulate known biology as well as genomic proximity effects.
a, Phenomics overview. Screening of the genetic perturbations produces images of cells from which features are extracted either using CellProfiler24 or neural networks. The feature vectors for each pair of perturbations are related using cosine similarity (ranging from −1 opposite (‘opp.’) to 1 similar (‘sim.’)) and visualized in heat maps. b, A heat map of genes with diverse functions. The rows and columns are clustered on the rxrx3 data. EGFR, epidermal growth factor receptor; TGFB, transforming growth factor-beta. c, Recall of annotated known relationships from three databases in the most extreme 10% of similarities (two sided). A random ranking of gene–gene pairs would give a baseline value of 0.1. d, Full-genome heat map where each row and column represent a gene assessed in both rxrx3 (above diagonal) and cpg0016 (below diagonal) studies. Ordering genes by chromosomal position reveals the proximity bias signal along the diagonal present in both datasets with the chromosome boundaries and centromeres clearly visible. e, A zoom-in on chromosome 8. f, Juxtaposition of chromosomes 5 and 19, where the pattern of proximity bias signal reflects a chromosomal rearrangement known to be present in U2OS cells (cpg0016 data) but not HUVEC (rxrx3 data). g, A bar plot of proximity bias metrics (Brunner–Munzel probabilities) for each chromosome arm for the rxrx3 dataset. The values above 0.5 indicate elevated intra-arm similarity, and all chromosome arms are significant with Bonferroni correction (one-sided P < 0.001). h, A bar plot of proximity bias metrics for each chromosome arm as in g for the cpg0016 dataset. All chromosome arms are significant with Bonferroni correction (one-sided P < 0.001). In all heat maps, each row and column represent a single gene with rxrx3 data shown above the diagonal, cpg0016 data below the diagonal. Only the 7,477 genes that are present in both datasets are shown. The solid lines represent chromosome boundaries and the dashed lines indicate centromeres.