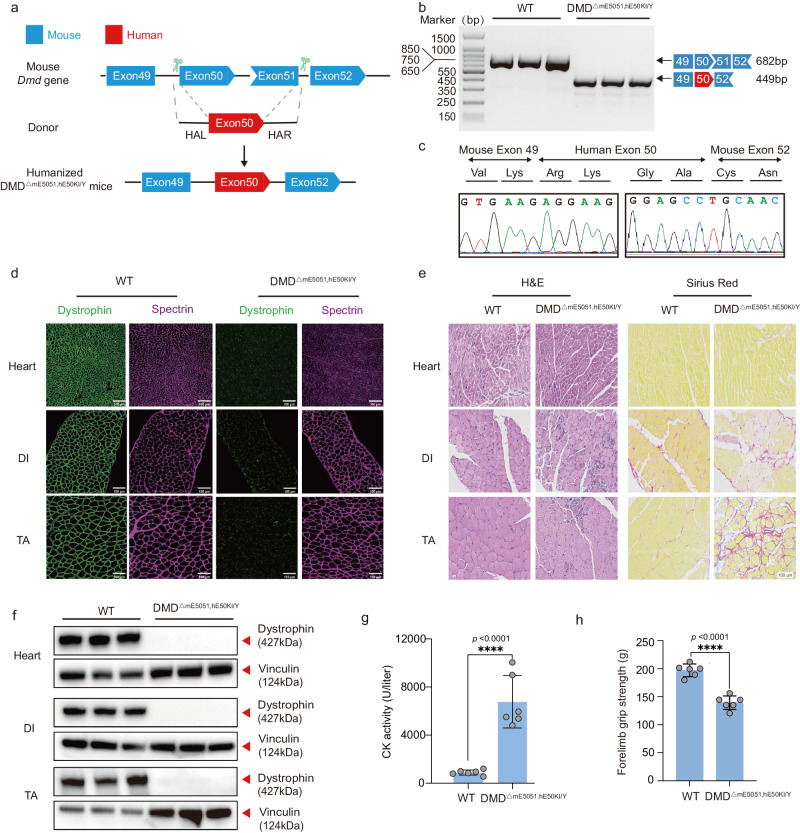
Fig. 1. Establishment and characterization of a humanized DMD mouse model.
a Strategy for generating humanized DMD mouse model. Shape and color of boxes of exons indicate reading frame. CRISPR-Cas9 editing using two sgRNAs flanking exon50 and exon51 was used to delete mouse Dmd exon 50 and exon51 (blue box), and replaced with human DMD exon 50 (red box) flanking 200 bp intron sequence. b RT-PCR products from muscle of DMDΔmE5051,KIhE50/Y mice were gel electrophoresis and sequenced (c) to validate exon knock in and deletion. d Dystrophin immunohistochemistry from indicated muscles of WT and DMDΔmE5051,KIhE50/Y mice. Dystrophin and spectrin are shown in green and mangenta, respectively. e Western blot confirming the absence of dystrophin in indicated muscle tissues. f Hematoxylin-eosin (H&E) and Sirius red staining of tibialis anterior (TA), diaphragm (DI), and heart muscle of wild-type (WT) and DMDΔmE5051,KIhE50/Y mice. g Serum creatine kinase (CK), a marker of muscle damage and membrane leakage, was measured in WT and DMDΔmE5051,KIhE50/Y mice. h WT and DMDΔmE5051,KIhE50/Y mice were subjected to forelimb grip strength testing to measure muscle performance. All mice were 8 weeks old at the time of the experiment. Data are presented as mean ± s.d (n = 6 independent biological replicates). Each dot represents an individual mouse. Significance is indicated by asterisk and determined using unpaired two-tailed Student’s t test. ****P < 0.0001. Scale bar, 100 μm. Source data are provided as a Source Data file.