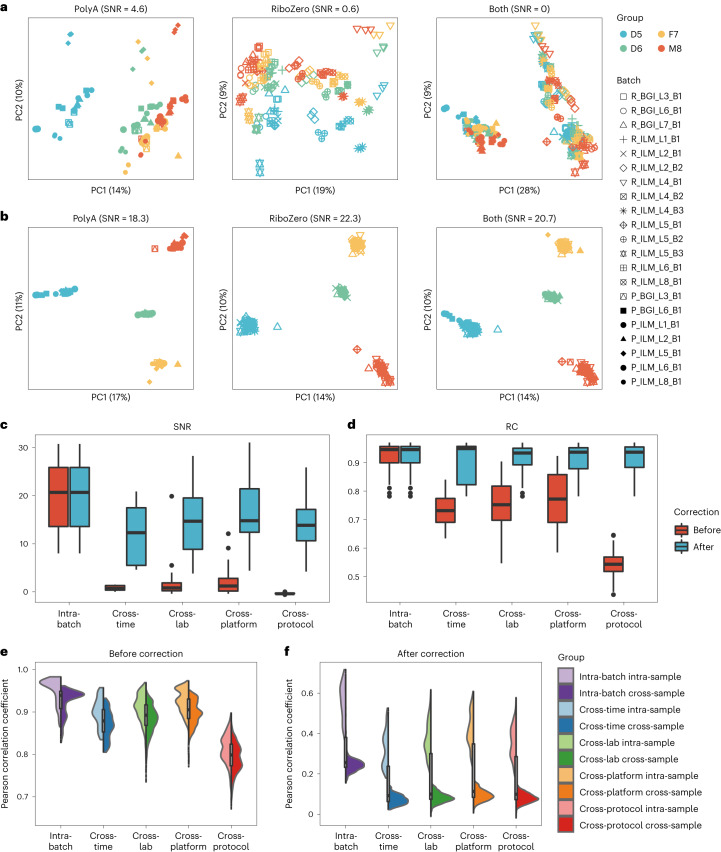
Fig. 4. Performance evaluation of cross-batch reproducibility.
a,b, Scatter plots of PCA on RNA-seq data before batch correction (a) and after correction (b) from replicates of the Quartet RNA reference materials (marked in colors) in the 21 batches (marked in shapes). Expressions in log2FPKM were used as before batch-correction datasets. Ratio-based expressions (which referred to converting expression profiles to gene-wise relative-scale profiles within each batch) were used to correct batch effects. Ratio-based expressions were obtained by subtracting log2FPKM by the mean of log2FPKM of the three replicates of D6 in the same batch. We used a multi-batch RNA-seq dataset, including 168 RNA-seq libraries from the RiboZero protocol and 84 RNA-seq libraries from the PolyA protocol. Plots were color-coded by sample groups and shaped by batches. c,d, Box plots of SNR values (c) and relative correlation with reference datasets (RC) values (d) for comparisons indicated at the x axis. When each batch of libraries was compared against each other, they could be classified into five different scenarios with increasing degree of differences, including intra-batch, cross-time, cross-laboratory, cross-platform of sequencing and cross-protocol levels. Intra-batch SNR values were calculated using 12 samples in the same batch, whereas SNR values of cross-batch were calculated by combining expression data from all combinations of two batches (n = 24). e,f, Violin plots of Pearson correlation coefficients based on expression profiles before (e) and after (f) batch correction for comparisons indicated at the x axis. D5, F7 and M8 samples were used to calculate pairwise correlations, whereas D6 samples were used as denominators for calculating ratio-based expressions for correcting batch effects. The number of combinations (n) used to derive statistics in c–f in each box were as follows: c: intra-batch, n = 21; cross-time, n = 7; cross-laboratory, n = 62; cross-platform, n = 43; cross-protocol, n = 98; d: intra-batch, n = 63; cross-time, n = 21; cross-laboratory, n = 186; cross-platform, n = 129; cross-protocol, n = 294; e and f: intra-batch intra-sample, n = 189; intra-batch cross-sample, n = 567; cross-time intra-sample, n = 189; cross-time cross-sample, n = 378; cross-laboratory intra-sample, n = 1,674; cross-laboratory cross-sample, n = 3,348; cross-protocol intra-sample, n = 1,161; cross-protocol cross-sample, n = 2,322; cross-platform intra-sample, n = 2,646; cross-platform cross-sample, n = 5,292.