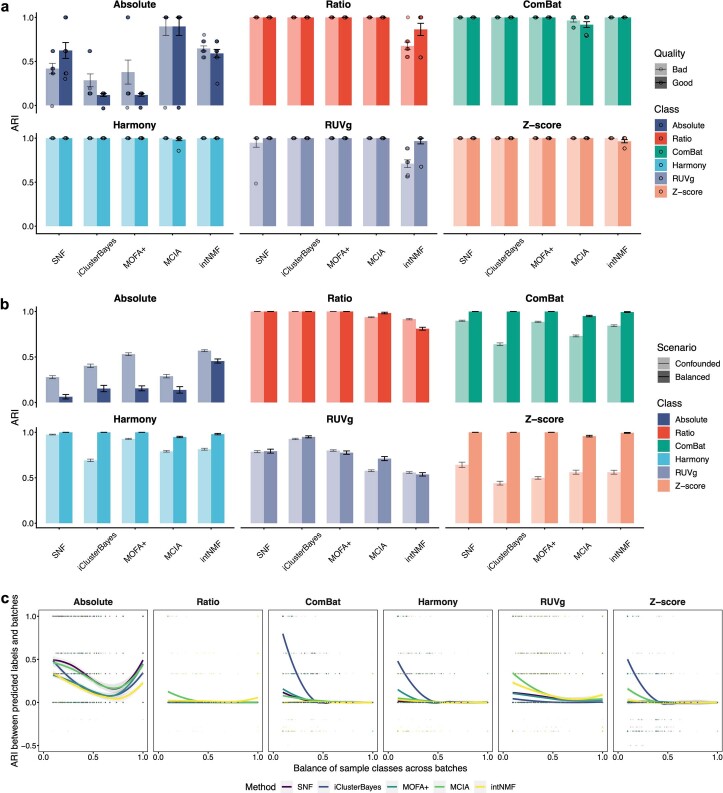
Extended Data Fig. 7. Vertical integration with different data preprocessing methods.
a,b, Bar plots of the Adjusted Rand Index (ARI) of vertically integrated multi-omics datasets of different quality datasets (a, Bad vs. Good) and different scenarios (b, Confounded vs. Balanced) at absolute level (Blue) and ratio level (Red) using SNF, iClusterBayes, MOFA + , MCIA, and intNMF. Data of each omics type were preprocessed by Absolute (no further processing on the normalized datasets), Ratio, ComBat, Harmony, RUVg, or Z-score for horizontal integration. The number of data sampling and integration instances (n) used to derive statistics were as follows: Bad, n = 10; Good, n = 10; Confounded, n = 200; Balanced, n = 100. Data are presented as mean values ± SD. c, Scatter plots between ARI between predicted labels and batches as well as the degree of sample class-batch balance with different data preprocessing methods. Each point represents an instance of data sampling and integration. The solid lines depict local regression fit of the data and shaded regions depict 95% confidence intervals.