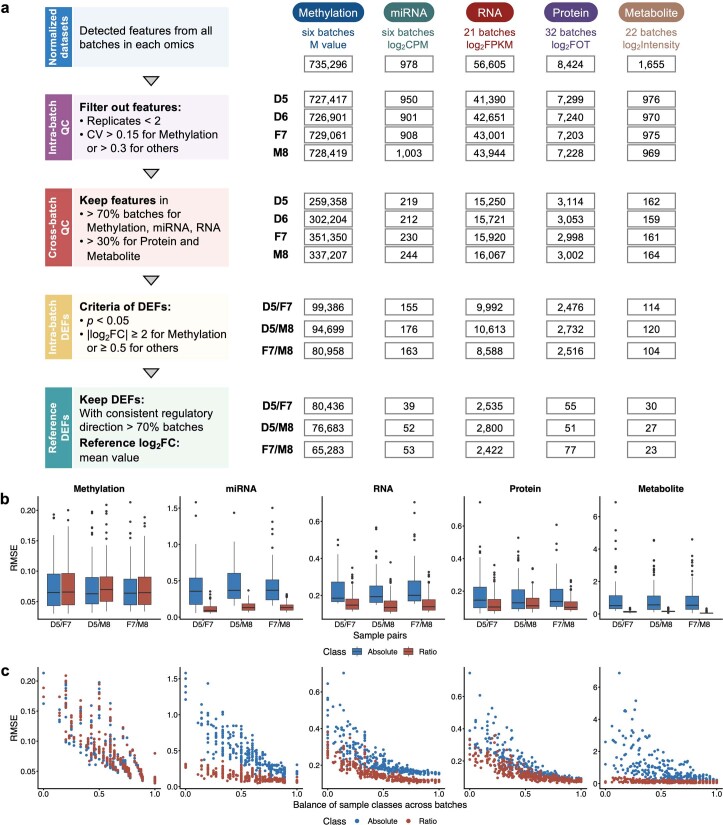
Extended Data Fig. 3. Ratio-based scaling promotes accurate identification of differentially expressed features.
a, Workflow for the construction of reference datasets of differentially expressed features. Reference datasets were constructed according to the following steps: (1) Identifying detectable multi-omics features and per-sample normalization. (2) Intra-batch quality control. Features that were not detectable or had low technical reproducibility were filtered out. (3) Cross-batch quality control. Features detectable in a sufficient number of batches were retained. (4) Calculating intra-batch differentially expressed features (DEFs) using t-test analysis. DEFs were classified as up- or down-regulated based on the positive or negative sign of the log2 fold change. (5) Voting based on the regulatory directionality (up or down) to screen the high confidence DEFs. b, Box plots of RMSE of the DEFs of horizontal integration data at absolute level (Blue) and ratio level (Red) based on the reference datasets. Data were sampled 100 times per pair of samples. The box plots display the distribution of data with the median represented by the line inside the box and the interquartile range (IQR) represented by the box. Whiskers extend to 1.5× the interquartile range. c, Scatter plots between RMSE when integrating at absolute (Blue) and ratio (Red) levels and the degrees of sample class-batch balance.