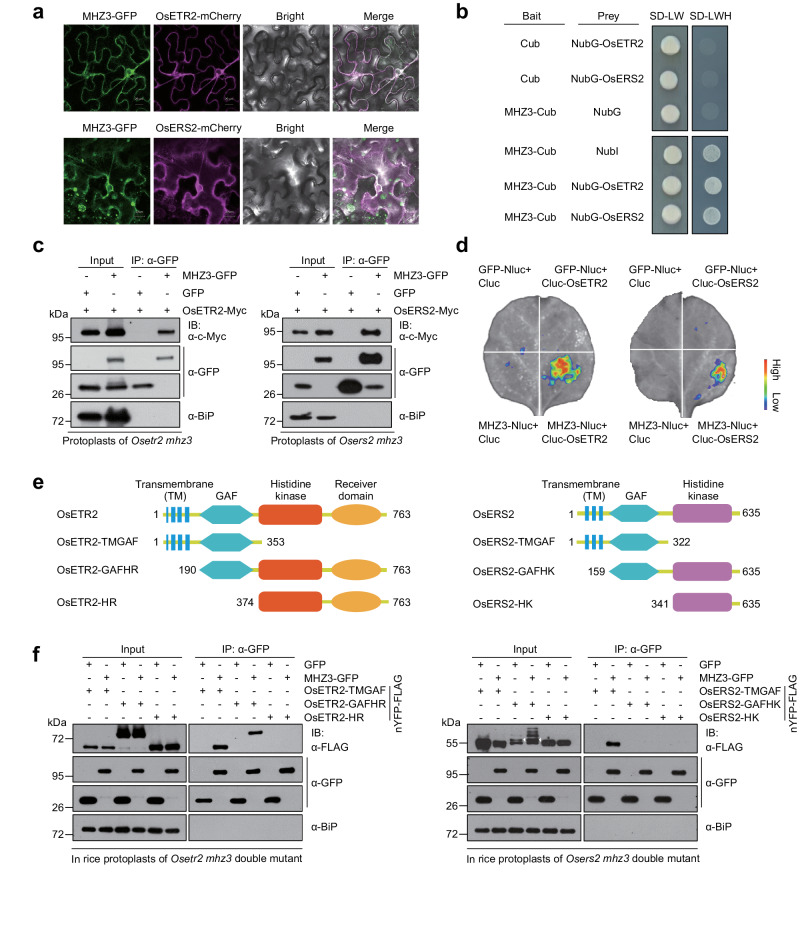
Fig. 3. MHZ3 physically interacts with OsETR2 and OsERS2.
a MHZ3 co-localizes with OsETR2 and OsERS2 in N. benthamiana leaves. Scale bars = 20 μm. b Split-ubiquitin membrane yeast two-hybrid assay for interaction of MHZ3 with OsETR2 and OsERS2. Cub, C-terminal half of the ubiquitin. NubI, N-terminal half of ubiquitin. NubG, Substitution of Ile-13 in wild-type NubI with glycine reduces Nub and Cub affinity. c Co-immunoprecipitation (Co-IP) assays for interaction of MHZ3 with OsETR2 and OsERS2. The indicated constructs were co-transformed into Osetr2 mhz3 or Osers2 mhz3 rice protoplasts. The GFP was used as a negative control. Total proteins were immunoprecipitated with GFP Trap and immunoblotted with anti-GFP, anti-c-Myc, and anti-BiP antibodies. IP: Immunoprecipitation. IB: Immunoblotting. d Luciferase complementation imaging assays for the interaction of MHZ3 with OsETR2 and OsERS2. Luciferase activity is visualized with artificial color from low (purple) to high (red). Nluc, N-terminal portion of firefly luciferase; Cluc, C-terminal portion of firefly luciferase. e Diagrams of full-length and truncated versions of OsETR2 and OsERS2 used in interaction domain mapping studies. GAF is short for cGMP-specific phosphodiesterases, Adenylyl cyclases, and FhlA. HR stands for histidine kinase domain plus receiver domain. HK stands for histidine kinase domain. f Co-IP assays for mapping interaction domains of OsETR2 and OsERS2 associated with MHZ3. The GFP was used as a negative control. The constructs were co-transformed into Osetr2 mhz3 or Osers2 mhz3 rice protoplasts. Total proteins were immunoprecipitated with GFP Trap and immunoblotted with anti-GFP, anti-FLAG, and anti-BiP antibodies. The triangle indicates the band for OsERS2-GAFHK-nYFP-FLAG. Two independent experiments were repeated with similar results. Uncropped blots in Source Data file.