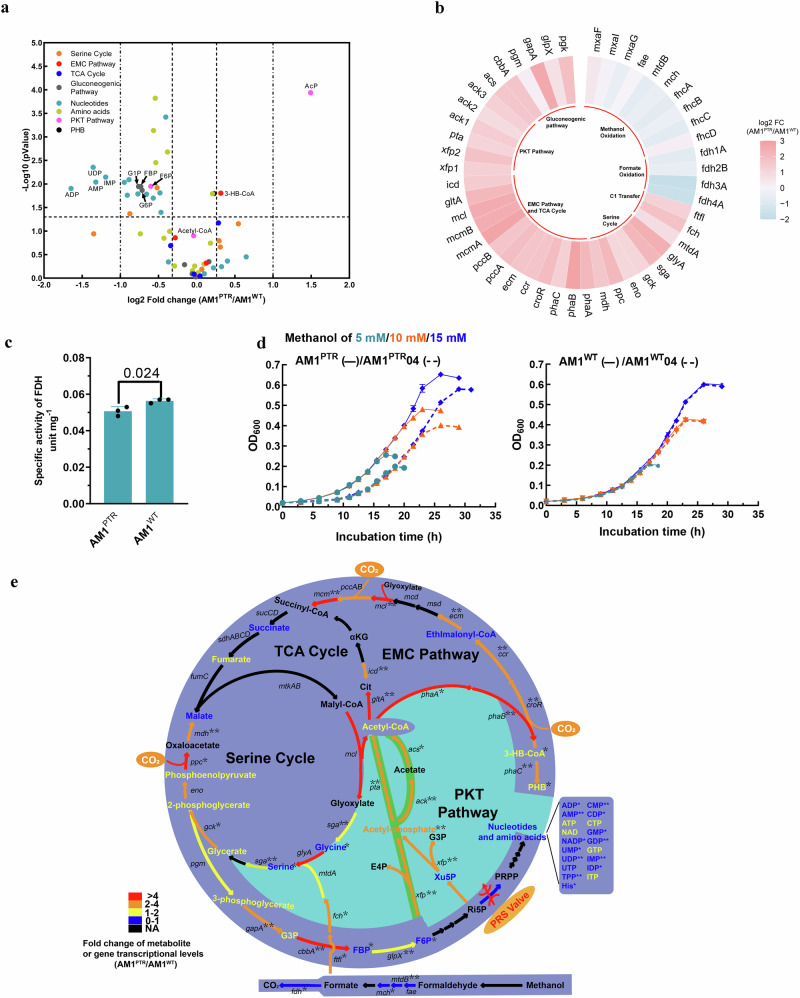
Fig. 3. PRS, as a metabolic valve, reallocates carbon flows through nucleotide anabolism and methylotrophic assimilation by up-regulating the PKT pathway.
a Comparison of the intermediates involved in the central methylotrophic metabolism and nucleotide synthesis of the AM1PTR and AM1WT strains grown on 10 mM methanol (n = 3 biologically independent samples.). b Transcriptional comparison of gene expression between the AM1PTR strain and AM1WT strain grown on 10 mM methanol (n = 3 biologically independent samples with five technical repeats). c Specific activities of FDH in the strains of AM1PTR and AM1WT grown on 10 mM methanol (n = 3 biologically independent samples with three technical repeats). d Knocking out the gene xfp2 (AM1PTR 04) decreased the growth advantage of the AM1PTR strain on low methanol concentrations of 5 mM, 10 mM, and 15 mM, while it did not affect the growth of the AM1WT strain (n = 5 biologically independent samples with three technical repeats.). e Schematic central methylotrophic network in the AM1PTR strain on low methanol concentration. Partially turning off the PRS valve decreased the allocation of limited methanol for nucleotide synthesis and up-regulated the PKT pathway to channel more methanol through the assimilation pathways. Reversible reactions are shown as double arrows. Fold changes of gene transcriptional levels in the AM1PTR strain versus the AM1WT are indicated by colored arrows. Fold changes of intermediate pools in the AM1PTR strain versus the AM1WT are indicated by colored metabolites. “NA” is abbreviated for “not analyzed”. F6P fructose-6-phosphate, G6P glucose-6-phosphate, G1P glucose-1-phosphate, Pyr pyruvate, FBP fructose-1,6-biphosphate, 3-HB-CoA 3-hydroxybutyryl-CoA, AcP acetyl-phosphate, PHB poly-β-hydroxybutyrate. Data are presented as mean ± SD. Significance was analyzed using a two-tailed t-test analysis. Source data are provided as a Source Data file.