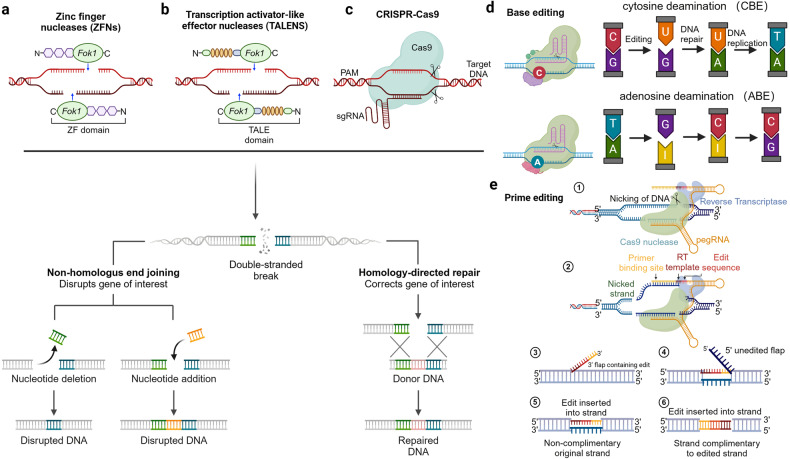
Fig. 6.
Various modes of genome editing. a ZFNs: By designing multiple zinc finger proteins to bind with specific DNA sequences, they guide the nuclease to cut the target gene, achieving precise gene editing through NHEJ and HDR. b TALENs: Specific transcription activator-like effector molecules are fused with nuclease domains. Two TALEN modules are necessary to bind to the target site, with a FokI nuclease fused to the DNA-binding domains, enabling precise gene editing through NHEJ and HDR. c CRISPR-Cas9: This system employs the Cas9 protein, guided by RNA molecules, to enable precise DNA cleavage at specific target sequences. It achieves accurate gene editing through NHEJ and HDR. d Base editing: an advanced form of Cas genome editing that enables the substitution of specific bases without inducing double-stranded breaks. This technique is categorized into two main types: C•G to T•A Base Editors (CBEs) and A•T to G•C Base Editors (ABEs). e Prime editing: The procedure involves constructing a prime editor complex, which includes a Cas domain, an RT (reverse transcriptase) domain, and a pegRNA. This complex searches for the DNA segment containing the target mutation and introduces a nick just in front of the mutation site. The nicked DNA strand then serves as a primer for DNA synthesis within the RT complex. The corrected, nicked strand is preferentially used over the original diseased strand, and the cell’s natural DNA repair mechanisms subsequently remove the diseased strand. This figure was created with Biorender.com