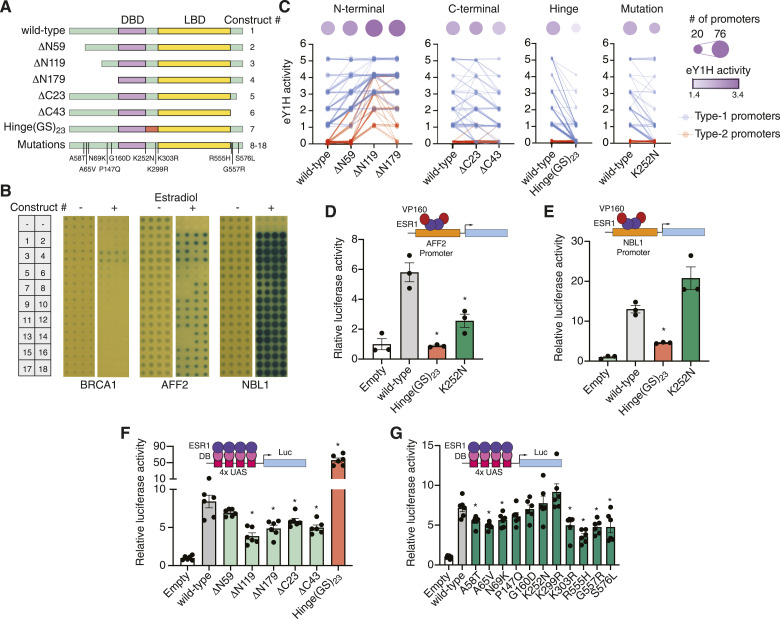
Figure 5. Role of ESR1 intrinsically disordered regions in DNA binding and transcriptional activity.
(A) Schematic of ESR1 constructs used. IDRs are indicated in green, DNA binding domain in purple, and ligand binding domain in yellow. (B) Examples of eY1H screens for binding of 18 different ESR1 constructs to the promoters of BRCA1, AFF2, and NBL1 in the presence or absence of 100 nM estradiol. (C) eY1H binding activity scored from 0 (no binding) to 5 (very strong binding) for different ESR1 constructs. Connected lines correspond to the same cancer gene promoter. Circle sizes indicate the number of bound cancer promoters, whereas color intensity indicates the average eY1H activity across bound promoters. Type 1 promoters (blue) are those that wild-type ESR1 binds; type 2 promoters (red) are those that wild-type ESR1 does not bind. (D, E) Luciferase assays in HEK293T cells where the promoters of AFF2 (D) or NBL1 (E) were cloned upstream of firefly luciferase. ESR1 constructs were fused to 10 copies of the VP16 activator domain. Experiments were conducted in biological triplicates. *P < 0.05. Statistical significance was determined by a two-tailed t test. (F, G) Mammalian one-hybrid assays measuring the transcriptional activity of different ESR1 constructs. ESR1 fusions with the Gal4 DNA binding domain (DB) are recruited to four copies of the Gal4 binding site (UAS) cloned upstream of firefly luciferase. Experiments were conducted in biological sextuplicates. *P < 0.05. Statistical significance was determined by a two-tailed t test.