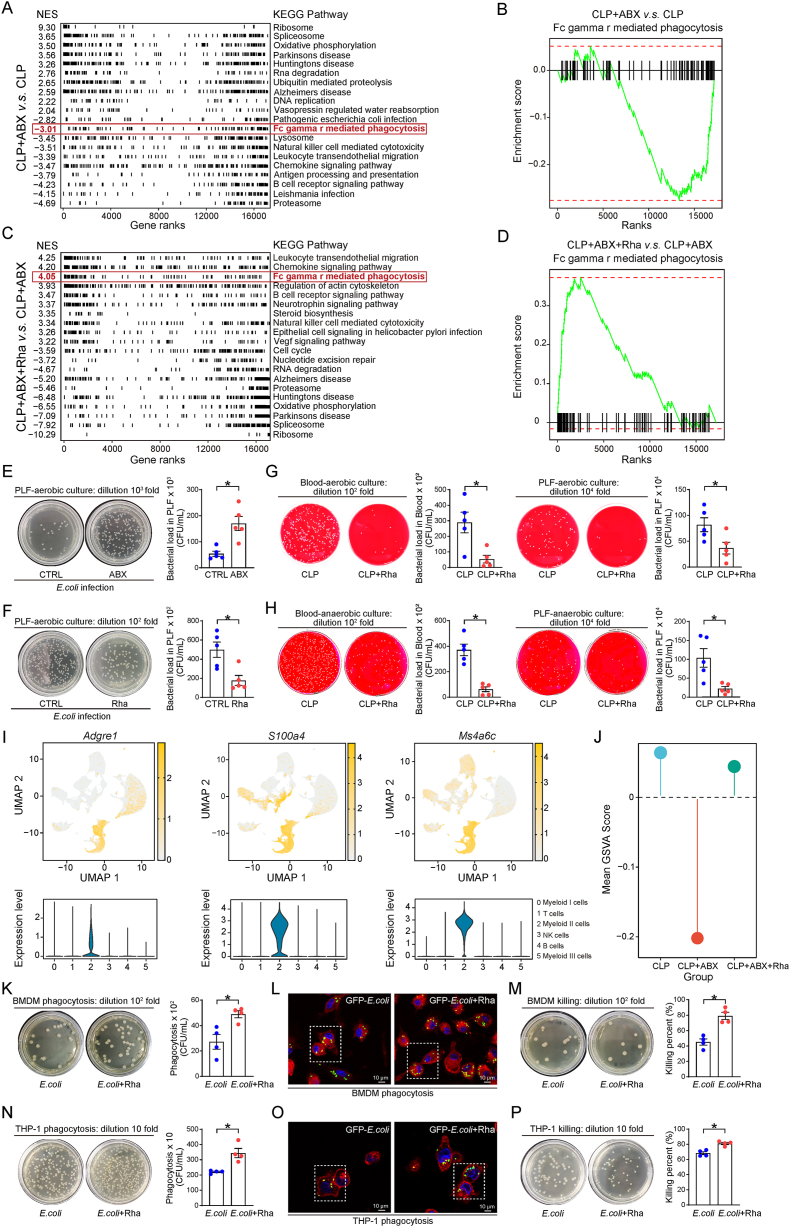
Figure 3.
Rhamnose decreased the bacterial load in sepsis mice by enhancing the bacterial clearance capacity of macrophages. (A) Pathway enrichment analysis for gene sets comparing peripheral blood mononuclear cells (PBMCs) from CLP + ABX group and CLP group. Mice were pretreated with PBS or ABX for three days followed by moderate CLP surgery, and then PBMCs were isolated from whole blood samples at 16 h for further single cell RNA sequencing (scRNA-seq). NES, normalized enrichment score, (n = 3/group). (B) Gene set enrichment analysis (GSEA) enrichment plot of FcγR-mediated phagocytosis signaling of total cells in CLP + ABX versus CLP group (n = 3/group). Experimental design as in Fig. 3A. (C) Pathway enrichment analysis for gene sets comparing PBMCs from CLP + ABX + Rha group and CLP + ABX group. Mice were pretreated with ABX for three days followed by rhamnose or PBS administration and then moderate CLP model was performed to induce sepsis, PBMCs were collected at 16 h after CLP model. NES, normalized enrichment score, (n = 3/group). (D) Gene set enrichment analysis (GSEA) enrichment plot of FcγR-mediated phagocytosis signaling of total cells in CLP + ABX + Rha versus CLP + ABX group (n = 3/group). Experimental design as in Fig. 3C. (E) Quantity of E. coli in the PLF at 12 h after injecting with E. coli in ABX and no-ABX pretreated mice (n = 5/group). (F) Quantity of E. coli in the PLF at 12 h after injecting with E. coli. Mice were orally administrated with PBS or rhamnose for 2 h followed by injection with E. coli (n = 5/group). (G, H) Quantification of bacterial colonies in the blood and PLF from the CLP mice in aerobic (G) or anaerobic (H) cultures. Mice were pretreated with PBS or rhamnose for 2 h followed by lethal CLP and tissue samples were collected at 12 h for the detection of bacterial load (n = 5/group). (I) According to macrophage marker genes (Adgre1, S100a4, Ms4a6c) to label single cells on a dimensional reduction plot. Violin plot evaluated the expression levels of macrophage marker genes in each cluster. (J) Gene set variation analysis (GSVA) enrichment lollipop chart of FcγR-mediated phagocytosis signaling in macrophages. The x-axes represent different group, and y-axes represent the average GSVA score in each group (n = 3/group). (K) Phagocytosis of E. coli in BMDMs. BMDMs were pretreated with rhamnose (100 μmol/L) or PBS for 18 h and then incubated with E. coli at 37 °C in 5% CO2 for 45 min with a multiplicity of infection (MOI) of 60 (n = 4/group). (L) Representative confocal images showing phagocytosis of GFP-E. coli by BMDMs. Experimental design as in Fig. 3K. Scar bar = 10 μm. (M) Percentage of E. coli killing by BMDMs in control (CTRL) and rhamnose (Rha) groups (n = 4/group). (N) Phagocytosis of E. coli in THP-1 derived macrophages (THP-1 cells). THP-1 cells were pretreated with rhamnose (100 μmol/L) or PBS for 18 h and then incubated with E. coli at 37 °C in 5% CO2 for 45 min with a MOI of 60 (n = 4/group). (O) Representative confocal images showing phagocytosis of GFP-E. coli by THP-1 cells. Experimental design as in Fig. 3N. Scar bar = 10 μm. (P) Percentage of E. coli killing by THP-1 cells in control (CTRL) and rhamnose (Rha) groups (n = 4/group). Data are expressed as mean ± SEM and P values were determined by a two-tailed unpaired Student's t-test. ∗P < 0.05 was considered statistically significant. n: indicates number of samples.