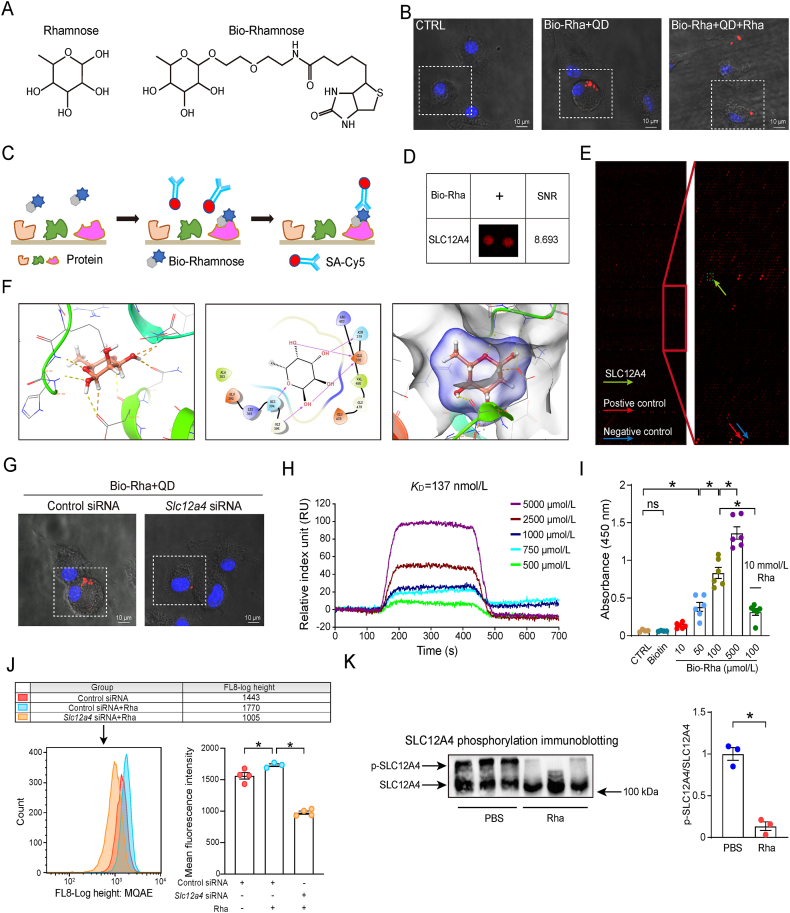
Figure 4.
Rhamnose interacts with SLC12A4 protein. (A) Chemical structures of rhamnose and biotinylated rhamnose (Bio-Rhamnose). (B) Representative confocal images of Bio-Rhamnose (Bio-Rha) binding to the surface of BMDMs. Confocal image of cells labeled with probes (red) and Hoechst 33342 stain (blue). Bio-rhamnose binding was blocked by free rhamnose (rhamnose + Bio-rhamnose). Scar bar = 10 μm. (C) A schematic diagram showing the experimental design for identifying rhamnose binding proteins using HuProt™20K Proteome Microarray Chip. (D) A magnified image showing interactions between Bio-rhamnose and SLC12A4 spots on the microarrays. Signal to noise ratio (SNR) is shown. (E) A representative image of HuProt™20K Proteome Microarray Chip showing positive (red arrow) and negative (blue arrow) spots, as well as spots for SLC12A4 (yellow spots). (F) Predicted binding modes of rhamnose targeting SLC12A4. Key residues are shown in red sticks, and hydrogen bonds are depicted by red arrows. (G) Representative confocal images of Bio-Rhamnose (Bio-Rha) binding to the surface of BMDMs pretreated with negative control or Slc12a4 siRNA. Cells were labeled with probes (red) and Hoechst 33342 stain (blue). Scar bar = 10 μm. (H) Direct interactions between rhamnose and SLC12A4 were determined by Surface plasmon resonance (SPR) analysis. (I) Rhamnose binding to SLC12A4 in ELISA. Recombinant hSLC12A4 was captured in wells pre-coated with anti-SLC12A4 antibodies. Bio-rhamnose at increasing concentrations and free rhamnose were added to perform the competition ELISA (n = 4–6/group). (J) Flow cytometry of MQAE intensity in BMDMs. Chloride ion could quench fluorescence signal, higher fluorescence intensity represents lower chloride ion concentration. Histogram of MQAE intensity (Mean Fluorescence Intensity) and the quantification (n = 3–4/group). (K) Western blot of phosphorylated SLC12A4 (p-SLC12A4) in BMDMs. Cells were treated with PBS or rhamnose for 5 min and the phosphorylation level of SLC12A4 was detected (n = 3/group). Data are expressed as mean ± SEM. One-way analysis of variance (ANOVA) analysis was used for three or more groups. Two groups were determined by a two-tailed unpaired Student's t-test, ∗P < 0.05 was considered statistically significant. ns: denotes not significant. n: indicates number of samples.