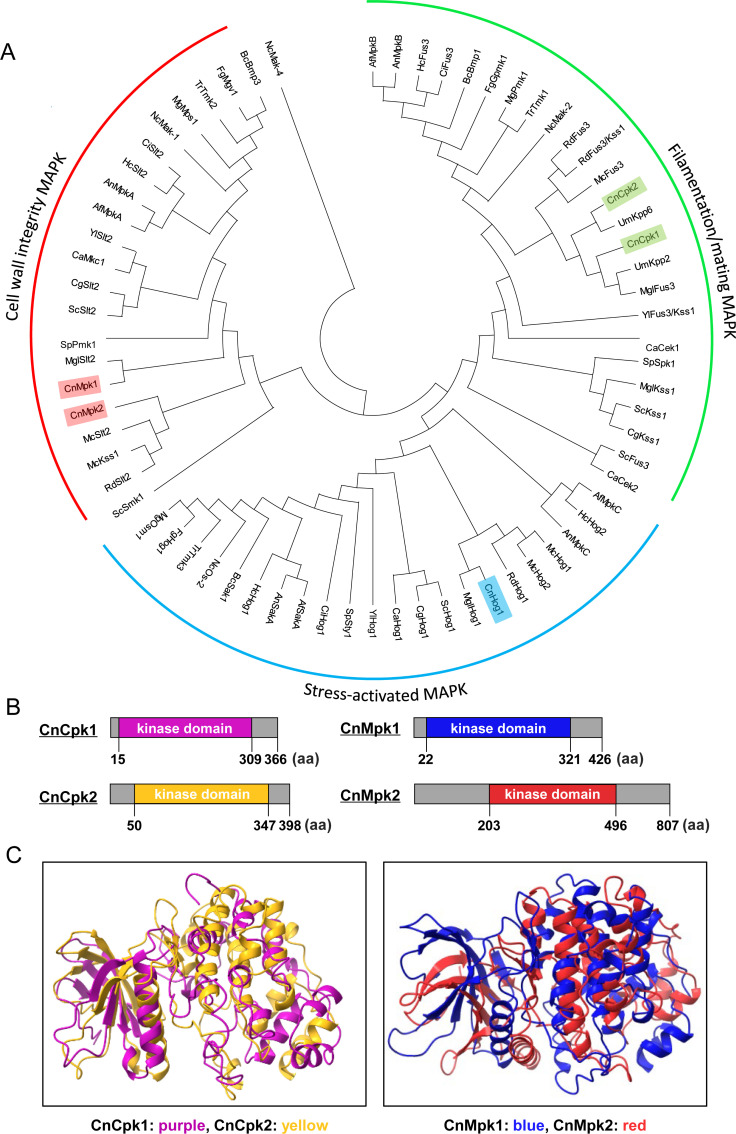
Fig 1.
Phylogenetic and structural analyses of Cpk1/Cpk2 and Mpk1/Mpk2 MAPK orthologs in fungi. (A) A phylogenetic tree illustrating MAPKs from 19 distinct fungal species is presented. The species abbreviations are as follows: Cn for C. neoformans; Sc for S. cerevisiae; Sp for Schizosaccharomyces pombe; Ca for Candida albicans; Cg for Candida glabrata; Fg for Fusarium graminearum; Mg for Magnaporthe grisea; Nc for Neurospora crassa; Af for Aspergillus fumigatus; An for Aspergillus niger; Bc for Botrytis cinerea; Um for Ustilago maydis; Yl for Yarrowia lipolytica; Rd for Rhizopus delemar; Hc for Histoplasma capsulatum; Mgl for Malassezia globosa; Mc for Mucor circinelloides; Tr for Trichoderma reesei; and Ci for Coccidioides immitis. (B and C) Illustrations depict the protein structures of the cryptococcal MAPKs: Cpk1, Cpk2, Mpk1, and Mpk2. Kinase domains for Cpk1 (in purple), Cpk2 (in yellow), Mpk1 (in blue), and Mpk2 (in red) were predicted by the InterPro domain analysis (https://www.ebi.ac.uk/interpro) and shown alongside their respective paralogs. Their kinase domain structures were predicted using AlphaFold2’s MMseqs2-based protein structure prediction method. Structural comparisons between Cpk1/Cpk2 and Mpk1/Mpk2 were visualized using the Mol*3D Viewer at the RCSB Protein Data Bank.