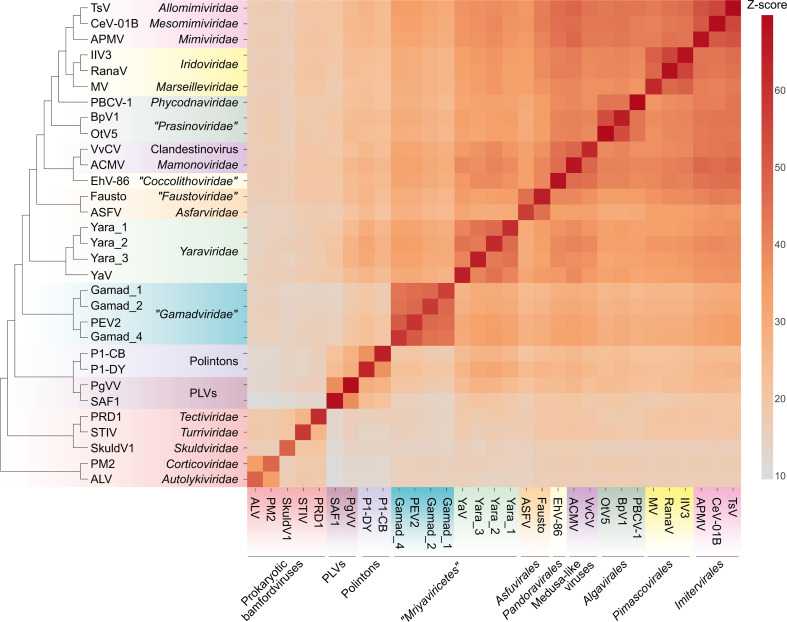
Fig 4.
Comparison of the predicted structures of mriyavirus major capsid proteins (MCPs) with structures of MCPs of other members of the kingdom Bamfordvirae. The heat map reflects the z-scores obtained in structural comparisons of the MCPs using Dali (color gradient shown to the right of the heat map). The dendrogram shows clustering of the MCPs by the z-scores. The abbreviations are as follows: ACMV, Acanthamoeba castellanii medusavirus (BBI30317); ALV, Vibrio phage 1.020.O._10N.222.48.A2 (AUR82054); APMV, Acanthamoeba polyphaga mimivirus (ADO18196.2); ASFV, African swine fever virus (PDB id: 6ku9); BpV1, Bathycoccus sp. RCC1105 virus BpV1 (YP_004061587); CeV-01B, Chrysochromulina ericina virus 01B (YP_009173446); EhV-86, Emiliania huxleyi virus 86 (YP_293839); Fausto, faustovirus (PDB id: 5j7o); Gamad_1, Ga0181388_1000587_17; Gamad_2, Ga0314846_0002864_7; Gamad_4, pleuro_group_Assembly_Contig_24_24; IIV3, invertebrate iridescent virus 3 (YP_654586); MV, Marseillevirus marseillevirus (YP_003407071); OtV5, Ostreococcus tauri virus 5 (YP_001648266); P1-CB, Polinton 1 of Caenorhabditis briggsae; P1-DY, Polinton 1 of Drosophila yakuba; PBCV-1, Paramecium bursaria chlorella virus 1 (PDB id: 5tip); PEV2, Pleurochrysis sp. endemic virus 2 (AUD57312); PLVs, polinton-like viruses; PM2, Pseudoalteromonas phage PM2 (PDB id: 2vvf); PRD1, Enterobacteria phage PRD1 (PDB id:1hx6); RanaV, Ranavirus maximus (YP_009272725); SkuldV1, Lokiarchaea virus SkuldV1 (UPO70972); STIV, Sulfolobus turreted icosahedral virus 1 (PDB id: 3j31); TsV, Tetraselmis virus 1 (YP_010783039); VvCV, Vermamoeba vermiformis clandestinovirus (QYA18424); Yara_1, Ga0364485_12008_8; Yara_2, Ga0466970_0005716_5; Yara_3, Yara_group_Contig_26_5; YaV, Yaravirus brasiliensis (QKE44414).