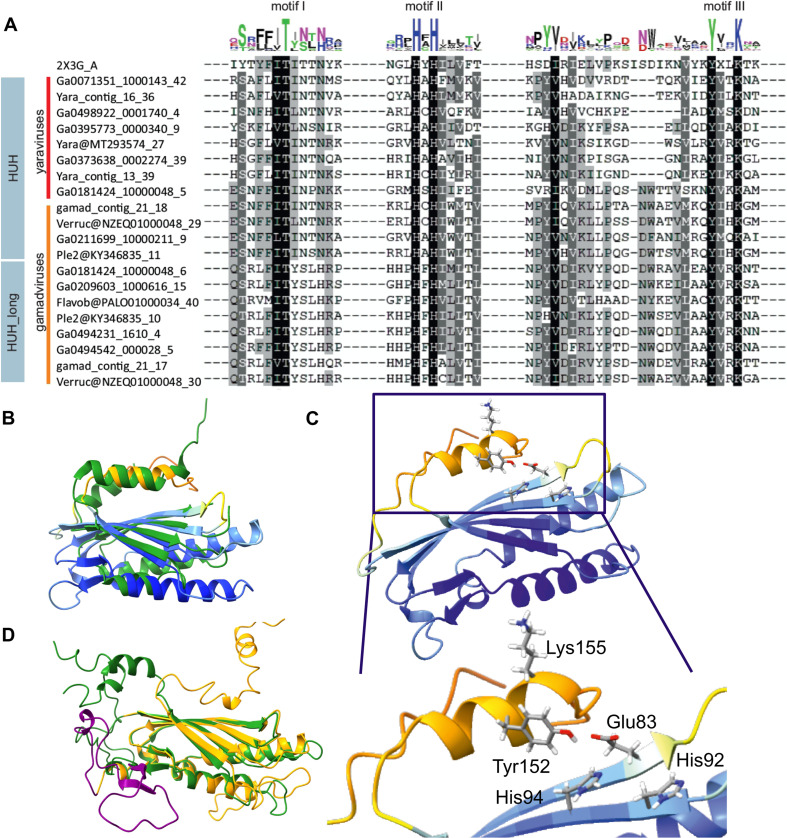
Fig 6.
Sequence and structure conservation in the HUH endonucleases of mriyaviruses. (A) Alignment of the sequence segments of the HUH superfamily endonucleases containing the characteristic motifs I–III. The N-terminal motif I consists of hydrophobic residues, motif II consists of HUH (H: Histidine, U: hydrophobic residue), and C-terminal motif III (Yx2-3K; Y: tyrosine, x: any residue, K: lysine, blue). (B) A representative predicted structure of a mriyavirus HUH endonuclease superposed with the crystal structure of protein ORF119 from Sulfolobus islandicus rod-shaped virus 1 (green, pdb 2X3G-A, z-score 7.7). Yaravirus HUH endonuclease (MT293574_27) colored by plddt score. (C) Configuration of the catalytic amino acid residues of motifs II and III in the predicted structure of the mriyavirus HUH endonuclease (yaravirus MT293574_27, colored by plddt score). (D) Superposition of the structural models of the two HUH endonuclease domains of gamadviruses (short: KY346835_11 [green, aa 31–224, aa 1–30 unstructured, clipped off for representation], long: KY346835_10 [orange, aa 1353–1574 with additional inserted loop shown in purple, aa 104–1450])].