Fig. 2 |. VAN-P assays accurately predict PDIM status during genetic manipulations and across different Mtb strains and lineages.
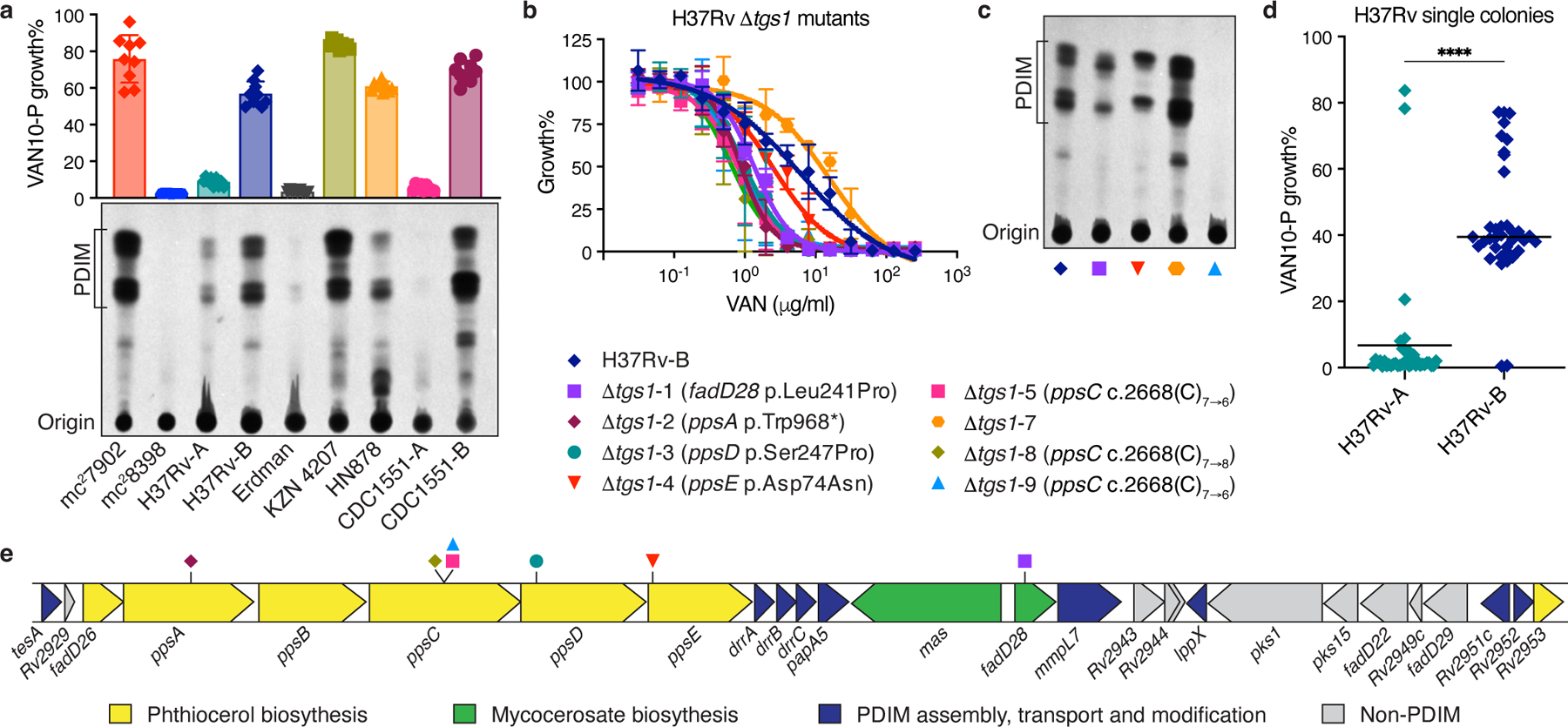
a, TLC lipid analysis and VAN10-P assays of different laboratory stocks of virulent Mtb strains alongside Mtb mc27902 and mc28398. Mean ± SD for n = 9 pairwise comparisons between triplicate wells. b, VAN-P MIC assays of eight Δtgs1 mutants and the parent H37Rv-B. Mean ± SD for n = 3 (Δtgs1-3, -5, -7 and -9) or n = 4 (H37Rv-B, Δtgs1-1, -2, -4, -8) biological replicates from two independent experiments. Mutations in PDIM biosynthetic genes are indicated in brackets in the legend. No mutations were detected in H37Rv-B or Δtgs1-7. c, TLC lipid analysis of four Δtgs1 mutants and H37Rv-B. Lipid extracts in (a) and (c) were run on the same TLC plate. d, VAN10-P screening of single colonies isolated from H37Rv-A (n = 38) and H37Rv-B (n = 37). Each colony was assayed in triplicate and data points represent mean VAN10-P growth%. Lines indicate the median. P < 0.0001; unpaired two-tailed Mann-Whitney test. e. Schematic showing the PDIM gene cluster and location of secondary PDIM mutations in Δtgs1 mutants.
