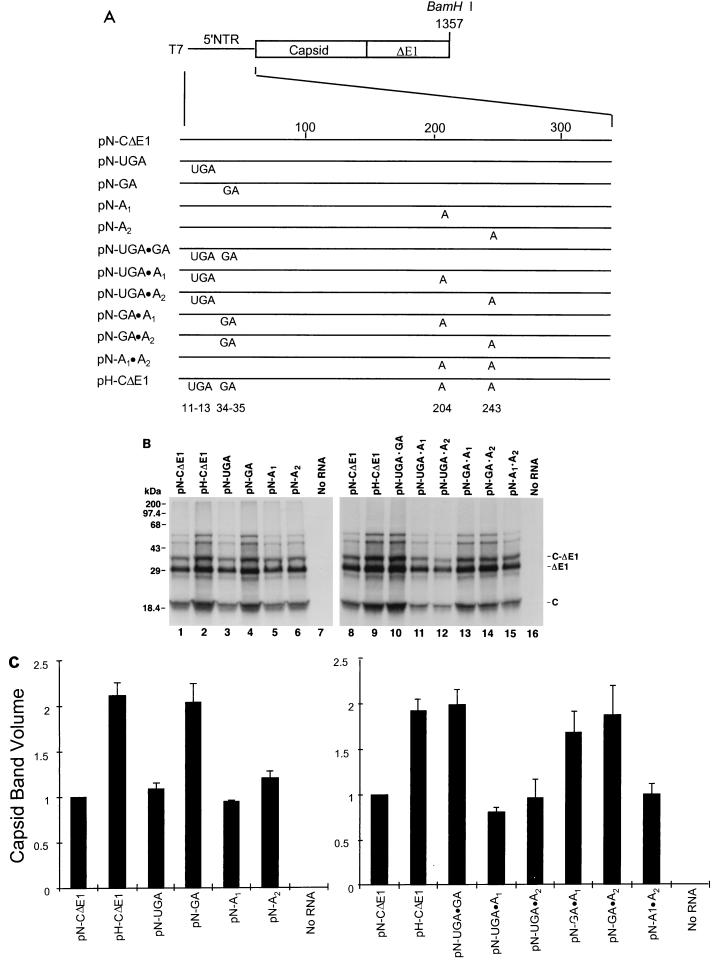
FIG. 3.
Genetic basis for the difference in translational activities of the HCV-N and HCV-H 5′NTRs. (A) Organization of T7 transcriptional units within chimeric constructs containing the unique nucleotide sequences of HCV-H placed individually and in combination within the background of the HCV-N 5′NTR and immediately downstream segment of the open reading frame encoding C-ΔE1 (pN-CΔE1). Protein-coding regions are shown as rectangles; noncoding RNA is shown as a solid line. Map positions of the base substitutions are shown at the bottom. (B) SDS-PAGE of products of translation from reticulocyte lysates programmed with RNAs transcribed from the plasmids depicted in panel A. The two panels represent results from separate experiments. The gel positions of the capsid (C), ΔE1, and nonprocessed C-ΔE1 protein products are shown at the right. (C) PhosphorImager analysis of capsid protein quantities produced in translation experiments such as those shown in panel B. The measured band volumes were normalized to that of reactions programmed with RNA from pN-CΔE1 (=1.0). The error bars represent the standard error calculated from the results of three separate experiments.