Figure 6.
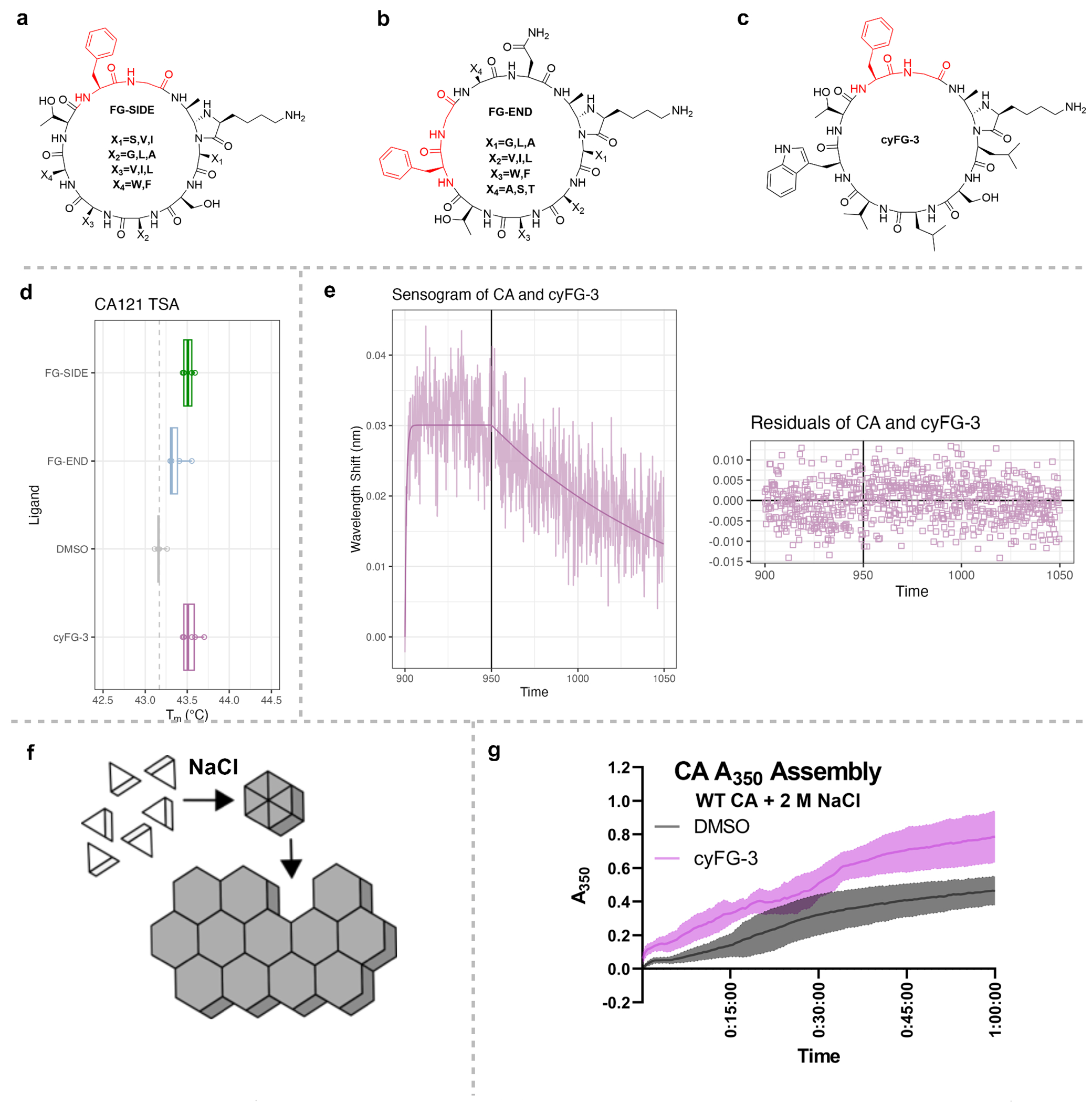
Example of library screening for cyclic peptides with phenylalanine-glycine (FG)-motifs that bind the HIV-1 Capsid protein (CA). Structures of (a) FG-SIDE (b) FG-END, (c) cyFG-3. (d) Average melting temperature (Tm) values as determined by Thermal Shift Assay (TSA) of CA hexamers incubated with the libraries of peptides containing the FG motif on the side (FG-SIDE) or end (FG-END), and the purified cyFG-3 from the FG-SIDE library. (e) Representative Biolayer Interferometry (BLI) sensograms of double background subtracted data and modeled fit for CA Hexamers with cyFG-3 at 75 μM (blue). Global KD = 0.963 ± 0.06 μM, kon = 8.90 ± 0.53 x103 M−1s−1, koff = 8.58 ± 0.136x10−3 s−1. Residuals of the model are shown below the plot as a difference between the modeled line and observed data. (f) Cartoon schematic of in vitro CA assembly. Purified CA monomers are clear in solution, but when a high concentration of NaCl increases the ionic strength of the solution, CA multimerizes into hexamers and large assemblies that turn the solution cloudy, which can be tracked with A350nm as a proxy of assembly. (g) Kinetics of in vitro CA assembly increase with pre-treatment with cyFG-3 (pink), as CA assembles faster than the control DMSO (grey). Average of 6 technical replicates (N = 3) with standard deviation shown.
