Abstract
The H4 subtype of avian influenza virus (AIV) exhibits a wide host range and is commonly found in migratory waterfowl. Recent studies have revealed that the H4N6 AIV can infect guinea pigs via aerosol transmission without prior adaptation. Additionally, the Q226L/G228S substitutions in the receptor-binding site have led to structural changes in globular head of H4 AIV, resulting in a configuration similar to that of pandemic H2N2 and H3N2 human influenza viruses. This article provides an updated review of the historical evolution, global distribution, adaptive mutations, receptor-binding preferences, and host range of H4 AIV. The insights presented herein will help in assessing the potential risk of future H4 AIV epidemics.
Key words: Avian influenza, Avian influenza virus, H4Nx, H4N6, receptor binding
INTRODUCTION
Avian influenza virus (AIV), a member of the Orthomyxoviridae family, can be classified into 2 distinct pathogenicity groups: highly pathogenic avian influenza virus (HPAIV) and low pathogenic avian influenza virus (LPAIV), based on their impact on poultry (Long, et al., 2019). HPAIV, particularly subtypes H5 and H7, have the capacity to precipitate severe disease outbreaks in poultry, leading to significant economic ramifications and potential public health concerns (Chatziprodromidou, et al., 2018; Luo, et al., 2021; Świętoń, et al., 2020). In contrast, LPAIV typically elicits mild or no clinical symptoms in poultry, with H4 AIV serving as a prime example. However, it has been observed that H4 AIV can co-circulate and undergo reassortment with other AIV subtypes, giving rise to novel viruses with increased pathogenicity (Liang, et al., 2016; Shi, et al., 2016; Quan, et al., 2018). Notably, the emergence of HPAIV H5N6, a reassortant virus with PB1 genes derived from H4N2, in Korea in 2016 exemplifies this phenomenon (Si, et al., 2017). Reassortment events involving H4 AIV and various hemagglutinin (HA) subtypes can indeed culminate in the emergence of novel HPAIV strains. Therefore, enhancing surveillance and monitoring efforts targeting H4 AIV assumes paramount importance, as it enables us to gain a deeper understanding of this virus and respond effectively to potential outbreaks.
The H4 subtype of AIV exhibits a wide range of host diversity and has been extensively documented in both wild and domestic avian species spanning multiple countries (Zhang, et al., 2012; Kang, et al., 2013; Yuan, et al., 2015; Hollander, et al., 2019). While waterfowl are considered the natural reservoir of H4 AIV, its presence has also been detected in various other avian species, such as parrots, quails, and turkeys, as well as in mammals including seals and swine (Matsuoka, et al., 1979; Kayali, et al., 2009; Wong, et al., 2014; Wu, et al., 2015; Xu, et al., 2020; Stallknecht, et al., 2022). Moreover, studies have demonstrated the ability of certain strains of H4 AIV to infect mice without prior adaptation, with respiratory droplets facilitating transmission in guinea pigs (Liang, et al., 2016). Swine, often referred to as the "mixing vessel" for AIV, can serve as a conduit for the reassortment of influenza viruses from diverse host species (Ito, et al., 1998; Skelton and Huber, 2022). Notably, the H4N6 swine influenza virus exhibits a preference for human α-2,6 sialic acid (SA) receptors, augmenting its potential to infect humans (Song, et al., 2017). There is no report of human infection with swine influenza H4, but it has been reported that humans have been infected with H4 subtype AIV. For example, in Lebanon, where migratory routes of wild birds converge, 3 backyard poultry farmers tested positive for H4 AIV through microneutralization assay, with titers of 1:10, 1:80, and 1:160, respectively (Kayali, et al., 2009; Kayali, et al., 2011). Given the close proximity between humans, swine, and poultry, the dissemination of AIVs capable of affecting both humans and animals has become increasingly prevalent, thereby emphasizing the significance of H4 AIV as a potential public health threat warranting attention. This review aims to provide an up-to-date overview of the surveillance and global distribution of H4 viruses among different hosts, as well as the adaptive mutations and receptor binding preferences of the H4 subtype of AIVs.
SURVEILLANCE OF H4 VIRUSES IN POULTRY AND WILD BIRDS
Domestic Poultry
Since its initial identification in a duck in 1956, the H4 strain of AIV has been detected across the globe. The phylogenetic analysis of the HA gene reveals 2 distinct lineages: North America and Eurasia (Donis, et al., 1989; Song, et al., 2017; Khan, et al., 2018). The surveillance of live poultry markets (LPMs) plays a pivotal role in evaluating the potential risk of AIV transmission to humans and in unveiling the origins of AIV outbreaks. From 2014 to 2021, a comprehensive sampling effort in China, encompassing 388,645 specimens collected from LPMs, slaughterhouses, farms, and backyard flocks, yielded a positive isolation rate of 0.09‰ for the H4 subtype (Bo, et al., 2022). Notably, all H4 isolates traced back to southern China, with the majority being identified in December.
In the province of Guangxi, China, between 2016 and 2019, a total of 7,567 samples sourced from LPMs resulted in the successful isolation of 974 strains of LPAIV, H3, H4, H6, and H9 were the most prevalent subtypes, with the H4 subtype constituting 5.03% of the isolates, among which mixed infections accounted for 8.62%. Interestingly, a significant portion of H4 isolates were recovered during the summer season. Among cases of mixed infection, the combination of H3 and H4 was the most prevalent, representing 33.33%, while H4 and H6 accounted for 11.90% (Luo, et al., 2021).
In the region of Sichuan, China, between 2014 and 2015, a total of 516 samples obtained from LPMs led to the isolation of 65 H4 AIV strains, predominantly aligning with the Eurasian lineage (Quan, et al., 2018). Furthermore, during the period from 2013 to 2014, 3,210 cloacal swab samples collected from domestic ducks in LPMs in Zhejiang, China, yielded 109 AIV strains, with 23 H4 AIV were isolated from domestic ducks. Genetic analysis confirmed that these H4 AIV strains shared genetic traits consistent with the Eurasian lineage (Wu, et al., 2015). In 2009, 3,787 oral and pharyngeal swab samples were obtained from ducks and chickens in LPMs in Shanghai, China, eight H4 AIVs were isolated from domestic ducks (Shi, et al., 2016).
Among 4,308 waterfowl samples collected from 4 LPMs in Bangladesh between 2007 and 2012, 191 were positive for AIV, of which eight strains of H4 subtype were identified (Khan, et al., 2018). However, in a surveillance study of AIV in poultry and wild birds in Korea from 2012 to 2014, it was found that H4 AIV was isolated more frequently in wild bird habitats than in poultry farms, but was not isolated in LPMs (Lee, et al., 2017). Notably, in LPMs in central Vietnam where disinfection interventions were conducted to reduce AIV transmission, two H4 AIVs were still isolated from environmental samples and from apparently healthy poultry oropharynx and cloaca (Chu, et al., 2016). LPMs are widely recognized as the primary source of AIV transmission and interspecies spread. Numerous studies have consistently shown the presence of H4 in LPMs, often coexisting with other AIV subtypes. Investigations conducted in Chinese LPMs have consistently revealed that the isolated H4 AIV strains belong to the Eurasian lineage, with no strains from the North American lineage detected. Nevertheless, reports regarding LPMs in other countries are limited, lacking a specific focus on the H4 subtype, and failing to delineate its lineage as Eurasian or North American.
Wild Birds
AIV is disseminated extensively by wild birds, making them invaluable surveillance species. Researchers collected a total of 8,594 fresh fecal samples from wild birds living in wetlands along the Yangtze River in central China's Hubei province from November to March, 2013 to 2017. Interestingly, only one strain of the H4 subtype AIV (A/Mallard/Hubei/chenhu_VI109/2015) (H4N8) was isolated from these samples (Wang, et al., 2021). Similarly, in China from 2015 to 2019, 10,910 fecal samples were collected from migratory birds in natural reserves along the East Asian-Australasian (EA) flyway (lakes and wetlands in Hunan, Hubei, and Jiangxi provinces, but no H4 AIV was isolated (Yao, et al., 2022). China exhibits a higher detection rate of H4 in LPMs compared to the habitats or stopover sites of migratory birds. From autumn 2019 to 2021, a total of 4,451 throat swabs, cloacal swabs and fecal samples were collected in the habitats of migratory birds in Shanghai. Notably, the highest prevalence of the H4 virus was observed. H4 was predominantly detected in the environment, with Anseriformes being the primary avian hosts. It is worth mentioning that all isolated H4N2 viruses belonged to the Eurasian lineages (Xu, et al., 2023).
In the Israel Hula Valley, a crucial waterbird habitat situated on a migratory bird route, Lublin et al. collected fecal samples from 53 wild mallards (Anas platyrhynchos) in 2018 and identified 5 strains as the H4N6 AIV (Lublin, et al., 2022). Monitoring surveys conducted from 1976 to 2015 in the Mississippi Migratory Flyway (MMF) and Central Migratory Flyway (CMF) indicated a positive rate of 11% for AIV, with H4 (26.2%) being one of the prominent subtypes (Diskin, et al., 2020). These strains were mainly concentrated in the summer and autumn seasons. Similarly, a survey conducted in Minnesota, USA from 2007 to 2018 indicated that H3 and H4 subtypes were prevalent in migratory mallards, particularly H3N8 and H4N6 (Hollander, et al., 2019; Stallknecht, et al., 2022).
A total of 291 H4 isolates from 22,229 fresh stool or cloacal swabs collected on the island on Oland Island, Sweden, between 2002 and 2010 were identified (Latorre-Margalef, et al., 2014). From 2002 to 2009, 289 H4 strains were isolated from 18,643 cloacal or fecal samples from migratory mallards in Sweden, with H4N6 being the most prevalent subtype, accounting for 79% of H4 isolates (Wille, et al., 2022). In India, from 2009 to 2011, 15 strains of the H4 subtype of AIV were isolated from 1,604 poultry samples (Pawar, et al., 2012). In South Korea's national survey from 2004 to 2010, a total of 30,881 samples were collected, with 52 isolated strains of H4 from wild bird fecal samples. The majority of these strains belonged to the Eurasian lineage, with a few belonging to the North American lineage (Kang, et al., 2013). Overall, the H4 subtype is prevalent among migratory mallards carrying AIV, thereby elevating the risk of poultry infection during their migratory journeys.
The H4 subtype of AIV is frequently detected in migratory birds, highlighting the importance of monitoring its potential to infect poultry during migration. A summary of H4 subtype AIV in different countries reported in the literature is shown in Table 1. Nonetheless, it remains uncertain whether the frequent mixed infections of H4 with other AIV subtypes, particularly H3+H4, in both migratory mallards and poultry markets contribute to the adaptation of the H4 subtype of AIV to poultry. Therefore, it is crucial to promptly comprehend the evolutionary dynamics of the H4 subtype of AIV. Such understanding is vital for comprehending its transmission capacity and predicting potential outbreaks in both livestock and humans.
Table 1.
Isolation positive rate of H4 subtype avian influenza virus in different countries reported in the literature.
| Country | Region | Period | Object | Sample | Total1 | H4 | Ref |
|---|---|---|---|---|---|---|---|
| China | Multiple regions | 2014-2021 | Environment | - | 388,645 | 0.09‰ | (Bo, et al., 2022) |
| Guangxi LPM | 2016-2019 | Domestic chickens, Ducks, Geese swab | Throat and cloacal swabs | 7,567 | 0.65% | (Luo, et al., 2021) | |
| Sichuan LPM | 2014–2015 | Domestic chicken and duck, Environment | Unspecified | 516 | 12.60% | (Quan, et al., 2018) | |
| Zhejiang LPM | 2013-2014 | Domestic ducks | Cloacal swabs | 3,210 | 0.72% | (Wu, et al., 2015) | |
| Shanghai LPM | 2009 | Domestic ducks and chickens | Oropharyngeal swabs | 3,787 | 0.21% | (Shi, et al., 2016) | |
| Chenhu Wetland,Honghu Wetland,Wanghu Wetland | 2013 - 2017 | wild bird | Fresh faeces | 8,594 | 0.01% | (Wang, et al., 2021) | |
| Shanghai habitat of migratory birds | 2019-2021 | Wild birds | Environmental samples (feces) and swab samples | 4,451 | 1.03% | (Xu, et al., 2023) | |
| Bangladesh | Multiple regions | 2007–2012 | Domestic waterfowl | Oropharyngeal, Cloacal, Fecal | 4,308 | 0.19% | (Khan, et al., 2018) |
| Vietnam | Thua Thien Hue province | 2014 | Domestic ducks, muscovy ducks, chickens and the environment | Oropharyngeal and cloacal swabs | 3,045 | 0.07% | (Chu, et al., 2016) |
| Israel | Hula valley | 2018 | Anas platyrhynchos | Fresh faeces | 53 | 9.43% | (Lublin, et al., 2022) |
| USA and Canada | Mississippi Migratory Flyway (MMF) and Central Migratory Flyway (CMF) | 1976-2015 | Migratory waterfowl | Cloacal, Oropharyngeal swabs, Blood | 77,969 | 2.83% | (Diskin, et al., 2020) |
| USA | Minnesota | 2014, 2015, 2017, 2018 in September | Anas platyrhynchos | Cloacal and oropharyngeal swabs | 2,972 | 5.69% | (Stallknecht, et al., 2022) |
| Minnesota | 2007-2016 | Waterfowl | Oropharyngeal and cloacal swabs | 13,228 | 4.27% | (Hollander, et al., 2019) | |
| Korea | Unspecified | 2004-2010 | Domestic ducks, Wild birds | Cloacal swabs or faeces | 30,881 | 0.17% | (Kang, et al., 2013) |
| India | West Bengal State | 2009–2011 | Environment, Migratory birds, Domestic ducks and poultry | Fecal droppings, Tracheal, cloacal swabs, Tissue, Serum | 5,722 | 0.26% | (Pawar, et al., 2012) |
| Sweden | Oland | 2002-2010 | Anas platyrhynchos | Fresh faeces or cloacal swabs | 22,229 | 1.31% | (Latorre-Margalef, et al., 2014) |
| Ottenby Bird Observatory | 2002-2009 | Anas platyrhynchos | Cloacal or faecal swabs | 18,643 | 1.55% | (Wille, et al., 2022) |
The total number of samples collected mentioned in the literature.
GLOBAL DISTRIBUTION OF H4Nx VIRUS
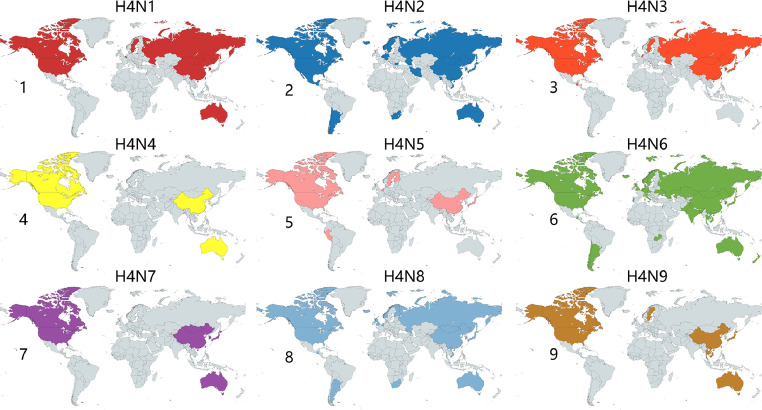
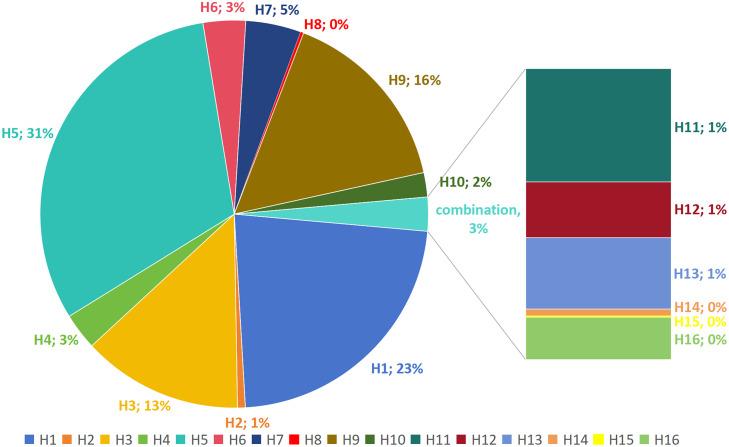
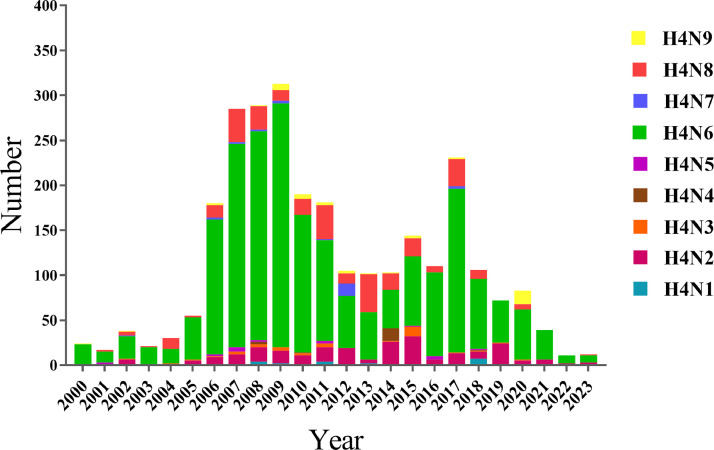
The Influenza Research Database (IRD), housed within the Bacterial and Viral Bioinformatics Resource Center (BV-BRC), has been harnessed to construct a comprehensive global distribution map of H4Nx AIV, as exemplified in Figure 1. The quantity and total count of distinct H4 subtypes isolated in various countries, across different time periods and species, have been compiled in Table 2. Additionally, the distribution of each HA subtype in the sequences deposited in GISAID between 2000 and 2023 is illustrated in Figure 2. Furthermore, temporal variations in the cumulative count of H4 AIV isolates have been presented in Figure 3. Notably, from 2000 to 2023, the predominant H4 subtype AIV isolates were H4N6, followed by H4N8 and H4N2.
Figure 1.
Global distribution of H4Nx influenza viruses. This figure displays the distribution of all currently available sequenced H4 viruses on the IRD (fludb.org). The maps represent the following: 1 = H4N1, 2 = H4N2, 3 = H4N3, 4 = H4N4, 5 = H4N5, 6 = H4N6, 7 = H4N7, 8 = H4N8, and 9 = H4N9. The graphics are visualized by www.mapchart.net.
Table 2.
All H4 hemagglutinin (HA) sequences currently available on Influenza Research Database- Sequence Search (fludb.org) with the corresponding years of detection and sampling source.
| Country | Period | Subtype | Species | No. of sequenced strains | Total |
|---|---|---|---|---|---|
| Argentina | 2011 | H4N2 | Duck (unspecified) | 1 | 5 |
| 2011-2016 | H4N6 | Teal, Duck(unspecified) | 3 | ||
| 2011 | H4N8 | Duck (unspecified) | 1 | ||
| Australia | 1994 | H4N2 | Duck (unspecified) | 1 | 20 |
| 1979 | H4N4 | Gray Teal | 3 | ||
| 1979-2001 | H4N6 | Gray Teal, Duck (unspecified) | 7 | ||
| 1980-2004 | H4N8 | Red-Necked Stint, Duck (unspecified) | 9 | ||
| Bangladesh | 2013-2017 | H4N6 | Duck (unspecified) | 8 | 8 |
| Barbados | 2004 | H4N3 | Blue-Winged Teal | 1 | 1 |
| Belgium | 2018 | H4N2 | Mallard | 1 | 4 |
| 2016-2018 | H4N6 | Mallard | 3 | ||
| Bulgaria | 2019-2010 | H4N2 | Duck (unspecified) | 4 | 4 |
| Canada | 1976-1998 | H4N1 | Waterfowl, Mallard, Mallard Duck | 3 | 317 |
| 1978-2009 | H4N2 | Mallard, Mallard Duck, Blue-Winged Teal, Pintail Duck | 9 | ||
| 1977-2007 | H4N3 | Mallard, Blue-Winged Teal | 3 | ||
| 1977-1983 | H4N4 | Mallard, Mallard Duck | 2 | ||
| 1990 | H4N5 | Blue-Winged Teal | 1 | ||
| 1976-2018 | H4N6 | Waterfowl, Duck, Mallard, Redhead Duck, Gadwall, Canvasback, Blue-Winged Teal, Northern Pintail, Pintail Duck, Swine, Green-Winged Teal, American Black Duck, Spot-Billed Duck, Environmental Sample | 274 | ||
| 2007 | H4N7 | Mallard | 1 | ||
| 1977-2016 | H4N8 | American Black Duck, Mallard, Blue-Winged Teal, Duck (unspecified), Pintail | 19 | ||
| 2009-2010 | H4N9 | Blue-Winged Teal, American Black Duck, Mallard | 5 | ||
| Chile | 2015-2016 | H4N2 | Yellow-Billed Teal, Mallard, Yellow-Billed Pintail | 3 | 6 |
| 2013 | H4N6 | Yellow-Billed Pintail | 3 | ||
| China | 1980-2009 | H4N1 | Swine, Duck (unspecified) | 2 | 134 |
| 1976-2014 | H4N2 | Duck, Goose, Chicken | 36 | ||
| 1977-2009 | H4N3 | Duck (unspecified) | 2 | ||
| 1978-2015 | H4N6 | Mallard, Duck (unspecified), Chicken, Goose | 75 | ||
| 1998 | H4N7 | Duck (unspecified) | 1 | ||
| 1998-2015 | H4N8 | Duck (unspecified), Chicken, Goose, Swine | 12 | ||
| 1977-2012 | H4N9 | Duck (unspecified), Environmental Sample | 6 | ||
| Czech Republic | 1956-2010 | H4N6 | Mallard, Duck (unspecified) | 9 | 9 |
| Georgia | 2011 | H4N2 | Garganey | 1 | 20 |
| 2016 | H4N6 | Mallard, Duck (unspecified) | 19 | ||
| Guatemala | 2011-2012 | H4N2 | Blue-Winged Teal, Northern Shoveler, American Wigeon | 9 | 16 |
| 2011 | H4N3 | Blue-Winged Teal | 3 | ||
| 2012 | H4N6 | Blue-Winged Teal | 1 | ||
| 2011-2013 | H4N8 | Blue-Winged Teal | 3 | ||
| Iceland | 2011 | H4N8 | Gull | 1 | 1 |
| India | 2009-2017 | H4N6 | Chicken, Mallard, Bird | 4 | 4 |
| Japan | 2004-2016 | H4N2 | Waterfowl, Avian, Mallard | 4 | 34 |
| 2006 | H4N3 | Duck (unspecified) | 1 | ||
| 2001 | H4N5 | Waterfowl, Duck | 2 | ||
| 1977-2017 | H4N6 | Budgerigar, Duck (unspecified), Avian, Waterfowl, Swan | 13 | ||
| 2012 | H4N7 | Stint | 2 | ||
| 2006 | H4N8 | Slaty-Backed Gull, Gull, Rufous-Necked Stint, Red-Necked Stint, | 9 | ||
| 2006-2017 | H4N9 | Waterfowl, Mallard | 3 | ||
| Kazakhstan | 2006 | H4N6 | Common Pochard, Common Coot | 2 | 2 |
| Mexico | 2011 | H4N2 | Environmental Sample | 1 | 1 |
| Mongolia | 2013 | H4N1 | Waterfowl | 1 | 42 |
| 2019 | H4N2 | Duck (unspecified) | 2 | ||
| 2007-2011 | H4N3 | Duck (unspecified), Waterfowl | 3 | ||
| 2009-2019 | H4N6 | Eurasian Wigeon, Waterfowl, Velvet Scoter, Duck (unspecified), Mallard, Teal, Shelduck, | 33 | ||
| 2002 | H4N7 | Duck (unspecified) | 1 | ||
| 2011-2012 | H4N8 | Duck (unspecified), Waterfowl | 2 | ||
| Netherlands | 2001-2012 | H4N2 | Mallard | 5 | 39 |
| 2014 | H4N3 | Mallard | 1 | ||
| 2006 | H4N5 | Black-Headed Gull | 1 | ||
| 1999-2017 | H4N6 | Mallard, Eurasian Wigeon, European Herring Gull, Mute Swan, Mallard, Bewick's Swan | 31 | ||
| 2006 | H4N8 | Common Eider | 1 | ||
| New Zealand | 1976-2004 | H4N6 | Mallard, Duck (unspecified) | 3 | 3 |
| Pakistan | 2010-2011 | H4N6 | Duck (unspecified), Chicken | 2 | 2 |
| Portugal | 2006 | H4N6 | Mallard | 1 | 1 |
| Russia | 2000-2012 | H4N6 | Sea Mammal, Mallard, Duck (unspecified), Shoveler, Gull, Shoveler, Muskrat, Pochard | 22 | 22 |
| South Africa | 2004-2013 | H4N8 | Red-Billed Teal, Cape Shoveler, Duck (unspecified) | 3 | 3 |
| South Korea | 2007 | H4N3 | Duck (unspecified) | 1 | 23 |
| 2005-2017 | H4N6 | Mallard, Duck (unspecified), Bird, Waterfowl | 20 | ||
| 2008 | H4N7 | Mallard | 1 | ||
| 2018 | H4N8 | Waterfowl | 1 | ||
| Sweden | 2006 | H4N1 | Mallard | 1 | 213 |
| 2002-2008 | H4N2 | Mallard | 17 | ||
| 2002-2009 | H4N3 | Mallard | 5 | ||
| 2006 | H4N5 | Mallard | 1 | ||
| 2002-2010 | H4N6 | Mallard, Dunlin | 187 | ||
| 2002-2009 | H4N9 | Mallard | 2 | ||
| Thailand | 2011-2012 | H4N6 | Duck (unspecified) | 2 | 2 |
| USA | 1977-2014 | H4N1 | Mallard, Northern Pintail | 7 | 1072 |
| 1976-2017 | H4N2 | American Wigeon, Mallard, Blue-Winged Teal, Green-Winged Teal, Turkey, Duck(unspecified), Northern Shoveler, American Black Duck, Northern Pintail, Quail, Turkey, Environmental Sample, Eurasian Teal | 80 | ||
| 1976-2017 | H4N3 | Green-Winged Teal, Pintail, Environmental Sample, Northern Shoveler, Mallard, Blue-Winged Teal, Northern Pintail | 8 | ||
| 2008-2014 | H4N4 | Northern Shoveler, Northern Pintail, Gull, | 15 | ||
| 1982-2019 | H4N5 | Sea Mammal, Mallard, Northern Pintail, Green-Winged Teal, Ruddy Turnstone | 11 | ||
| 1974-2011 | H4N6 | American Wigeon, Green-Winged Teal, Unknown, Duck, Mallard, Blue-Winged Teal, Northern Shoveler, American Black Duck, Environmental Sample, Northern Pintail, Cinnamon Teal, Gadwall, Swine, Eurasian Teal, American Green-Winged Teal, Goose, Bufflehead, Gull, Ring-Necked Duck, Snipe, Ruddy Turnstone, Red Knot, Shorebird, Waterfowl, Pintail, White-Winged Scoter | 739 | ||
| 2007-2017 | H4N7 | American Wigeon, Northern Pintail, Mallard, Gull | 4 | ||
| 1975-2018 | H4N8 | Chicken, Mallard, Blue-Winged Teal, Duck, Black Duck, Turkey, Bufflehead, Least Sandpiper, Northern Pintail, Ring-Necked Duck, Grebe, American Green-Winged Teal, Environmental Sample, Common Goldeneye, Ruddy Turnstone, Greater Scaup | 196 | ||
| 1986-2017 | H4N9 | Mallard, Ruddy Turnstone, American Black Duck, Blue-Winged Teal | 12 | ||
| United Kingdom | 2014 | H4N6 | Chicken | 2 | 2 |
| Viet Nam | 2009-2015 | H4N6 | Duck (unspecified), Muscovy Duck | 4 | 4 |
| Zambia | 2008 | H4N6 | Goose | 1 | 1 |
Figure 2.
Proportion of AIV subtypes isolated globally from 2000 to 2023. The data is based on the GISAID database and includes all hemagglutinin (HA) subtypes of influenza viruses (except human). The combination includes H11-H16, accounting for a total of 3%, with detailed proportions shown in the bar chart.
Figure 3.
Temporal Evolution of Total H4 Avian Influenza Viruses (AIVs) Worldwide from 2000 to 2023. Each color represents a different neuraminidase (NA) subtype, and the length of the cylinder corresponds to the quantity of H4Nx.
PREVALENCE OF H4 IN DIFFERENT HOSTS
Waterfowl
The risk of AIV infection in poultry is increased due to frequent contact between wild birds and waterfowl, as waterfowl can serve as silent carriers of various subtypes of AIV (Liu, et al., 2020). Transmission of AIV among waterfowl primarily occurs through fecal-oral routes and water-related activities such as preening infected feathers and cloacal drinking (Wille, et al., 2018). Among waterfowl, dabbling ducks, particularly Anas platyrhynchos (Olsen, et al., 2006), are the primary carriers of AIV. The H4 subtype of AIV has been identified as predominant in Anas platyrhynchos (Hollander, et al., 2019; Stallknecht, et al., 2022; Wille, et al., 2022), suggesting its potential role in the dissemination of H4 subtype AIV and the provision of internal genes for highly pathogenic strains. For instance, the H5N6 highly pathogenic AIV, isolated from migratory birds in South Korea, represents a novel reassortant virus incorporating genetic material from at least 3 different subtypes (H5N6, H4N2, and H1N1), with the H4N2 subtype contributing the PB1 gene (Si, et al., 2017). While the evolutionary rate of the 3 internal genes (PA, PB1, PB2) of H4 AIV in wild birds is slower compared to that of H5N1 in poultry (Fourment and Holmes, 2015), it is crucial to monitor the evolutionary dynamics of H4 in a timely manner as it is indeed evolving in wild birds.
Chicken
Studies have indicated that the adaptability of H4 AIV in chickens is not as strong as in ducks (Shi, et al., 2016; Luo, et al., 2021). In 2012, an H4N2 virus (A/Quail/California/D113023808/2012) was isolated from quail brain tissue, which exhibited a preference for α-2,6 type receptors and contained a polybasic CS motif 322PEKRRTR/G329 (Wong, et al., 2014). This virus showed low pathogenicity in chickens. However, H4 has the potential to enhance its adaptability in chickens through reassortment with H5N1 subtypes, regardless of the T327K/R substitution in the CS motif (Veits, et al., 2012; Gischke, et al., 2020). It was observed that low pathogenic LPAIV A/Mallard/Germany/1240/1/2007 (H4N6) acquired a polybasic HA cleavage site (CS) after co-transfection with the highly pathogenic strain A/Swan/Germany/R65/2006 (H5N1). Subsequently, the LPAIV H4 variant, featuring multiple basic CS, exhibited a level of pathogenicity in chickens comparable to that of the HPAIV H5 strain, with a chicken intravenous pathogenicity index (IVPI) of 2.79. The reassortant strain exhibited the HA-F281I substitution following passage in chicken embryos (Veits, et al., 2012). Reassortment of the H4 subtype with other highly pathogenic HPAIV is a feasible process. While clinical symptoms resulting from LPAIV infections in poultry are generally milder compared to those caused by HPAI, they are not entirely absent. Typical manifestations of LPAIV in poultry may include mild respiratory issues, slight reductions in egg production, and a low mortality rate, usually below 5%. It is crucial to recognize that more severe clinical outcomes can occur, especially in cases of coinfections with other pathogens, such as bacteria. The H4N6 subtype of AIV was identified in the UK within a laying flock exhibiting escalating daily mortality rates and decreased egg production over a 5-d period. Postmortem examinations of deceased birds revealed bronchitis, pulmonary congestion, air sacculitis, secondary peritonitis, and ovarian degeneration (Liu, et al., 2003; Reid, et al., 2018). In experiments conducted by Morales et al., most instances of H4 infection did not induce significant clinical disease or visible lesions, although one strain of H4N9 resulted in the death of a turkey and moderate respiratory symptoms and depression in a chicken (Morales, et al., 2009). While H4 AIV currently poses a limited threat to chickens, it is important to acknowledge that the H4 subtype of AIV may serve as a genetic reservoir for highly pathogenic AIV through genetic reassortment and adaptive mutations.
Swine Influenza
The H4N6 subtype of swine influenza virus (A/Swine/Ontario/01911-1/99 and A/Swine/Ontario/01911-2/99) was first identified in naturally infected swine in Canada in 1999, with affected pigs displaying symptoms like coughing, respiratory distress, weight loss. The internal genes (PB2, M, NS) of this strain originated from H9 and LPAIV H7 subtypes, all sourced from avian hosts (Karasin, et al., 2000a; Karasin, et al., 2000b). In China, H4 AIV was also detected in pigs (Ninomiya, et al., 2002). In 2009, the H4N1 subtype (A/Swine/HuBei/06/2009) was isolated from a severely ill pig with respiratory syndrome in China, with all viral genes exhibiting avian characteristics and sharing high sequence similarities (Hu, et al., 2012). Another strain, H4N8 (A/Swine/Guangdong/K4/2011(H4N8)), was found in swine in Guangdong, China, with its NP gene originating from H5N1 AIV (Su, et al., 2012). Notably, two H4 swine influenza virus strains from China featured Q226 and G228 in the receptor binding site (RBS) of the HA protein, displaying a preference for α-2,3 SA receptors.
In 2015, an H4N6 strain (A/swine/Missouri/A01727926/2015) was isolated from swine in the United States, causing flu-like symptoms but showing limited transmissibility among swine (Abente, et al., 2017). Hayashi et al. identified avian H4N6 virus with antigenic similarity to A/swine/Missouri/A01727926/2015, leading to the development of an inactivated whole-virus vaccine against A/swan/Hokkaido/481102/2017 (H4N6) (Hayashi et al., 2020). Amino acid sequence analysis revealed that the HA of Canadian and American swine isolates contained the L226/S228 configuration, which has an affinity for human α-2,6 SA receptors. Presently, there are no documented cases of human infection with the H4 subtype of swine influenza virus.
Human
Two cases of human infection with avian H4 virus have been documented. In a study conducted between July and September 2010, 200 volunteers with poultry contact and 50 without were recruited, and blood samples were successfully obtained from 248 participants. Utilizing a microneutralization assay, the results indicated that backyard poultry growers may have been previously infected with H4 and H11 AIV, suggesting the potential for H4 and H11 influenza viruses to cross the species barrier and infect humans (Kayali, et al., 2011). Additionally, a US study carried out between March 2007 and April 2008 involving 95 professional turkey workers (57 turkey growers, 38 turkey meat processing plant workers) and 82 unexposed controls revealed that backyard and free-range turkey farmers had been infected with AIV of H4, H5, H6, H9, and H10 subtypes. This was determined through the evaluation of 170 blood samples using microneutralization and hemagglutination inhibition tests (Kayali, et al., 2010) .
Ferrets, Mice, and Guinea Pigs
Ferrets, mice, and guinea pigs are commonly used animal models for studying influenza virus pathogenicity and transmission in humans (Belser, et al., 2011; Bouvier, 2015). Recent studies have indicated that the H4 subtype of AIV may have the ability to directly infect mammalian hosts. H4 viruses can replicate in the respiratory organs of infected mice without requiring pre-adaptation (Kang, et al., 2013; Wu, et al., 2015; Liang, et al., 2016). A study conducted in Korea found that the replication titer of H4 virus in quails and mice was relatively higher compared to ducks (Kang, et al., 2013). Additionally, Bui VN et al. isolated an H4N8 virus from shorebirds that caused mortality in mice (Bui, et al., 2012). Experiments conducted in guinea pigs have revealed that certain H4 viruses can be transmitted through direct contact, while others can be transmitted via respiratory droplets, albeit with limited efficiency (Liang, et al., 2016). Consequently, it is crucial to undertake additional research on the droplet and aerosol transmission capabilities of H4 viruses, with the utilization of ferret models being particularly valuable in assessing the risk of H4 virus transmission to humans.
ADAPTATION AND MUTATIONS IN H4Nx VIRUSES
Hemagglutinin (HA)
Several glycosylation sites (GS) near the apex of the HA spike can influence the binding affinity between HA molecules and host SA (Luo, 2012; Pralow, et al., 2021). In the context of BV-BRC, the HA protein of the H4 subtype exhibits 5 potential N-GS, namely 19NYT, 35NGT, 179NLT, 311NIS, and 499NGT. In-depth analysis reveals that while the GS of H4 remains relatively conserved, the amino acid at position HA-315 can undergo an Isoleucine to Valine mutation. However, the significance of this mutation remains to be investigated.
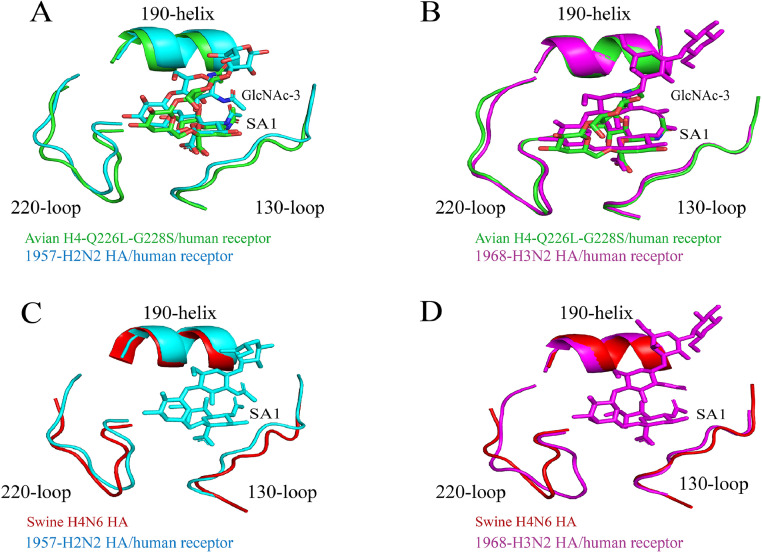
The RBS of H4 is highly conserved and comprises the 130-loop, 190-helix, and 220-loop. The asparagine residue at position 193 is believed to play a pivotal role in H4′s binding to the human receptor α-2,6 SA (Abente, et al., 2017; Liang, et al., 2016). The dominant combination of Q226/G228 is observed among the 2,252 stored H4 HA sequences in the IRD database, indicating that most H4 viruses exhibit a preference for binding to SA receptors with 2,3-linkages in avian species. The L226/S228 combination has only been identified in 3 strains isolated from naturally infected swine and has the ability to enhance the infectivity of H4 virus in primary swine and human respiratory epithelial cells (Bateman, et al., 2008; Liang, et al., 2016). Nevertheless, a single Q226L mutation alone is insufficient to facilitate the spread and sustained infection of H4 virus in swine (Abente, et al., 2017). Song et al. demonstrated that the HA-Q226L/G228S mutation combination, when present in avian species, alters the conformation of the avian RBS by increasing the distance between the 130-loop and 220-loop. Consequently, avian H4 viruses acquire a preference for binding to human receptors and exhibit receptor-binding characteristics similar to those of the pandemic H2N2 and H3N2 viruses (Song, et al., 2017). Conversely, the swine isolate H4N6 (A/Swine/Ontario/01911-1/99) carrying the HA-226L-228S mutation demonstrates notable distinctions when compared to H2N2 and H3N2 viruses (Figure 4). It is important to note that this mutation combination has not been observed in avian species thus far. Following continuous passage in mice, H4N6 virus displayed substitutions of L331I and G453R in the HA2 region of the HA protein. Although these mutations do not affect the receptor binding preference of H4N6 virus, the combination of L331I and G453R enhances the pathogenicity of H4N6 in mice (Xu, et al., 2020). Further investigations are required to determine whether the substitution of this mutation combination in other species would increase the virulence of H4 virus.
Figure 4.
Structural binding of human receptor analogues to avian H4-Q226L-G228S mutants and swine H4N6 HA. The HAs from the 1957 pandemic (A/Singapore/1/1957 H2N2, PDB: 2WR7) are shown in cyan, the 1968 human H3N2 (A/Aichi/2/1968 H3N2, PDB: 2YPG) in magenta, the HA-Q226L-G228S site-directed mutant (Avian HA-Q226L-G228S A/duck/Czechoslovakia/1956 H4N6, PDB: 5XLD) in green, and the Swine H4N6 (A/swine/Ontario/01911-1/99 H4N6, PDB: 5XL2) in red. The complex structures were compared for the following: (A) Avian H4-Q226L-G228S with 1957-H2N2 HAs bound to the human receptor analog, (B) Avian H4-Q226L-G228S with 1968-H3N2 HAs bound to the human receptor analog, (C) Swine H4N6 with 1957-H2N2 HAs bound to the human receptor analog, and (D) Swine H4N6 with 1968-H3N2 HAs bound to the human receptor analog.
Neuraminidase (NA)
AIVs have evolved adaptations to thrive in terrestrial poultry populations by undergoing deletions in the NA stalk region. In 2009, no deletion was detected in the H4 NA isolated from LPMs in Shanghai (Shi, et al., 2016). And, 2010-2011, waterfowl habitat separation from Mexico H4N2 strain in NA area did not appear the lack of handle (Ornelas-Eusebio, et al., 2015). On the other hand, Yuan et al. identified H4N6 AIV in ducks from China in 2012, which did exhibit the deletion in the NA protein stalk region. Interestingly, these H4N6 strains had not yet developed the E119V and R292K substitutions that provide resistance to NA inhibitors (Yuan, et al., 2015). However, the current literature on H4 AIV is limited, and the downloading of H4 subtype NA sequences from the GISAID database revealed few NA stem deletions. Additionally, residues E119 and R292 were found to be conserved.
Internal Genes
The PB1-F2 protein, a pro-apoptotic protein found in influenza A viruses, has been identified as playing a pathogenic role in mouse models. Even a single amino acid substitution in PB1-F2 can lead to an increase in viral virulence. For example, the N66S amino acid change has been associated with enhanced virus pathogenicity (Zamarin, et al., 2006; Conenello, et al., 2007; Kamal, et al., 2017). During a survey of LPMs in China in 2009, a strain with the PB1-N66S mutation was discovered (Shi, et al., 2016). Additionally, an H4N6 strain with this substitution was isolated during monitoring efforts in India between 2016 and 2017 (Pawar, et al., 2021). In 2006, an H4N8 strain was isolated from seagulls, and although its PB1-F2 protein consisted of 101 amino acids, the N66S mutation was not detected. The study did, however, identify a K482R substitution in its PB2 protein, but the significance of this substitution has yet to be demonstrated (Bui, et al., 2012; Cheng, et al., 2014; Jiao, et al., 2014; Liu, et al., 2018). Investigations conducted in Korea and China have not detected the E627K substitution in the H4 PB2 protein, nor have they found the K526R, A588V, and D701N substitutions (Kang, et al., 2013; Song, et al., 2014; Liang, et al., 2016; Liu, et al., 2018; Quan, et al., 2018). Nonetheless, the mouse-adapted strain BJ21-MA exhibited E158K and/or E627K mutations in the PB2 protein, which significantly increased polymerase activity, virus replication, and virulence (Xu, et al., 2020). Several H4 strains have exhibited mutations in the M2 protein, such as L26F and V27I, which confer resistance to the anti-influenza drugs amantadine and rimantadine (Kang, et al., 2013; Liang, et al., 2016; Shi, et al., 2016). In 2012, an E92D substitution was discovered in the NS1 gene of a duck-origin H4N6 strain isolated in China, and this substitution is known to be associated with severe pathology in mammals (Yuan, et al., 2015). Therefore, continuous monitoring of H4 is crucial as it possesses the ability to enhance its pathogenicity or alter its receptor binding preference through substitutions at specific sites. While other AIV subtypes have shown relatively few substitutions that enhance pathogenicity, certain H4 strains have exhibited resistance to influenza drugs, which may impede future prevention and control efforts. Consequently, prioritizing the development of effective H4 vaccines is of utmost importance in order to prevent and control H4 mutations and reassortment. We have summarized a table of amino acid substitutions mentioned above that affect the virulence and receptor preference of H4 subtype avian influenza viruses in Table 3.
Table 3.
Mutations impacting the virulence/receptor preference of H4Nx AIVs.
| Protein | Strain | Mutation1 | Host2 | Effect | Refs |
|---|---|---|---|---|---|
| HA | A/swine/Ontario/01911-1/99(H4N6) | Q226L/G228S | Swine | Increased affinity for swine and human-type receptor | (Karasin, et al., 2000a; Karasin, et al., 2000b) |
| A/swine/Ontario/01911-2/99(H4N6) | Q226L/G228S | Swine | Increased affinity for swine and human-type receptor | (Karasin, et al., 2000a; Karasin, et al., 2000b) | |
| A/swine/Missouri/A01727926/2015(H4N6) | Q226L | Swine | Only minor damage to the lungs | (Abente, et al., 2017) | |
| A/mallard/Beijing/21-MA/2011(H4N6)a | L331I/G453R | Mice | When occurring concurrently, creates increased virulence | (Xu, et al., 2020) | |
| PB1-F2 | A/duck/Shanghai/421-2/2009 (H4N6) | N66S | Domestic duck | Creates increased virulence | (Shi, et al., 2016) |
| A/migratory bird/India/1722760/2017 (H4N6) | N66S | Migratory bird | Creates increased virulence | (Pawar, et al., 2021) | |
| A/slaty-backed gull/Japan/6KS0185/2006(H4N8) | K482R | Slaty-backed gull | Unstudied | (Bui, et al., 2012) | |
| PB2 | A/mallard/Beijing/21-MA/2011(H4N6)a | E158K and/or E627K | Mice | Enhanced replication and virulence | (Xu, et al., 2020) |
| M2 | A/duck/Guangdong/S1469/10(H4N2),A/duck/Hubei/S2213/12(H4N2),A/duck/Hubei/S2227/12(H4N2) | V27I | Duck | Resistance - related substitution of anti-influenza drugs amantadine and rimantadine | (Liang, et al., 2016) |
| A/duck/Anhui/S2193/12(H4N6) | S31N | Duck | |||
| A/chicken/Guangdong/S1010/10(H4N8) | L26F/V27I | Chicken | |||
| A/duck/Shanghai/MH-2/2009 (H4N6), A/duck/Shanghai/44-2/2009 (H4N6), A/duck/Shanghai/46-2/2009 (H4N6), A/ duck/Shanghai/67-2/2009 (H4N6) | V27I | Domestic duck | The amantadine resistance marker | (Shi, et al., 2016) |
H3 numbering used.
Host the virus was isolated from in the relevant study: (e.g., A/mallard/Beijing/21-MA/2011(H4N6), the mutation was identified when serially passaged in mice, not in the original avian isolate).
RECEPTOR BINDING CHARACTERISTICS OF H4Nx VIRUS
Human influenza viruses have a preference for binding to α-2,6 SA receptors in a cis-configuration, whereas AIV primarily show a preference for α-2,3 SA receptors in a thin and straight trans configuration (Bateman, et al., 2008; Long, et al., 2019). Notably, α-2,3-sialylated glycans are abundant in human alveolar cells, while α-2,6 linkage is dominant in epithelial cells of the nasal mucosa, paranasal sinuses, pharynx, terminal bronchiole walls, and respiratory bronchiole walls (Liu, et al., 2018). Despite their higher affinity for α-2,3-linked glycans, certain H4 AIV strains can also recognize α-2,6 SA receptors (Liang, et al., 2016; Abente, et al., 2017). This contrasts with Song et al.'s findings, where they demonstrated that avian H4 viruses only bind to α-2,3-linked glycans. Song et al. further showed that the HA-Q226L substitution is crucial for avian H4 viruses to bind to human receptors, while the HA-G228S/A substitutions enable dual receptor binding ability (Song, et al., 2017).
It is important to note that the HA-Q226L substitution has not been observed in avian H4 viruses in the BV-BRC database. However, strains with the HA-G228S substitution (A/northern pintail/Interior Alaska/10BM07242R0/2010(H4N6)) and 2 strains with the HA-G228A substitution (A/mallard/Minnesota/sg-00824/2008(mixed)) and (A/American green-winged teal/Interior Alaska/10BM08226R0/2010(H4N6)) have been identified. Nevertheless, there has been no research conducted to analyze the ability of these substituted strains to bind to human receptors, as well as their pathogenicity or transmissibility. While human infection with H4 AIV currently does not show clinical signs, it is crucial to consider the potential risk of cross-species transmission with H4 AIV strains, especially as they may acquire substitutions that are compatible with human SA receptors.
CONCLUSIONS AND PERSPECTIVES
In summary, the study of the H4 subtype of AIVs remains limited, despite its wide host range, spanning from wild waterfowl to marine animals, poultry, and even mammals. While it is predominant among wild waterfowl, its adaptability in chickens is compromised. However, in LPMs or backyard farms, it can intermingle with other AIV subtypes, leading to reassortment and increased fitness in chickens. The ability of certain H4 viruses to transmit via respiratory droplets in guinea pigs poses a potential threat to human health. It is essential to enhance the understanding of the receptor binding characteristics of avian H4 viruses through the use of more sensitive methodologies.
Additionally, the non-compulsory reporting of H4 subtypes has resulted in a lack of recent submissions of H4 sequences to the IRD database, making its latest evolutionary dynamics elusive. Recent human fatalities caused by H3N8 infections in China have been reported, with mixed infections of H4 and H3 subtypes being most prevalent in LPMs. Further investigation is needed to explore the relationship between these subtypes. Notably, the prevalence of H4 AIV varies significantly along different bird migratory routes. Therefore, it is crucial to enhance surveillance of H4 AIV, elucidating the circulating subtypes and their evolutionary patterns. Such efforts can help assess the risk of cross-species transmission of H4 AIV.
DISCLOSURES
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was funded by GuangDong Basic and Applied Basic Research Foundation (Grant No. 2022A1515012462 and 2020A1515010116), Research Project of Education Department of Guangdong Province, China (2022KTSCX124), Laboratory open innovation fund project of Foshan University (KFCX2024-B26) and Guangzhou Zengcheng District Entrepreneurship Leading Team Project (202101001).
REFERENCES
- Abente E.J., Gauger P.C., Walia R.R., Rajao D.S., Zhang J., Harmon K.M., Killian M.L., Vincent A.L. Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the Midwestern United States. Virology. 2017;511:56–65. doi: 10.1016/j.virol.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A.C., Busch M.G., Karasin A.I., Bovin N., Olsen C.W. Amino acid 226 in the hemagglutinin of H4N6 influenza virus determines binding affinity for alpha2,6-linked sialic acid and infectivity levels in primary swine and human respiratory epithelial cells. J. Virol. 2008;82:8204–8209. doi: 10.1128/JVI.00718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser J.A., Katz J.M., Tumpey T.M. The ferret as a model organism to study influenza A virus infection. Dis. Model Mech. 2011;4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo H., Zhang Y., Dong J., Li X.Y., Liu J., Tan M., Zhao X., Wang D.Y. [Distribution and gene characteristics of H3, H4 and H6 subtypes of low pathogenic avian influenza viruses in environment related avian influenza viruses during 2014-2021 in China] Zhonghua Yu Fang Yi Xue Za Zhi. 2022;56:1549–1553. doi: 10.3760/cma.j.cn112150-20220810-00803. [DOI] [PubMed] [Google Scholar]
- Bouvier N.M. Animal models for influenza virus transmission studies: a historical perspective. Curr. Opin. Virol. 2015;13:101–108. doi: 10.1016/j.coviro.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui V.N., Ogawa H., Xininigen K.K., Matsuo K., Awad S.S.A., Minoungou G.L., Yoden S., Haneda H., Ngo L.H., Tamaki S., Yamamoto Y., Nakamura K., Saito K., Watanabe Y., Runstadler J., Huettman F., Happ G.M., Imai K. H4N8 subtype avian influenza virus isolated from shorebirds contains a unique PB1 gene and causes severe respiratory disease in mice. Virology. 2012;423:77–88. doi: 10.1016/j.virol.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatziprodromidou I.P., Arvanitidou M., Guitian J., Apostolou T., Vantarakis G., Vantarakis A. Global avian influenza outbreaks 2010–2016: a systematic review of their distribution, avian species and virus subtype. Syst. Rev. 2018;7:17. doi: 10.1186/s13643-018-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K., Yu Z., Chai H., Sun W., Xin Y., Zhang Q., Huang J., Zhang K., Li X., Yang S., Wang T., Zheng X., Wang H., Qin C., Qian J., Chen H., Hua Y., Gao Y., Xia X. PB2-E627K and PA-T97I substitutions enhance polymerase activity and confer a virulent phenotype to an H6N1 avian influenza virus in mice. Virology. 2014;468-470:207–213. doi: 10.1016/j.virol.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Chu D.H., Okamatsu M., Matsuno K., Hiono T., Ogasawara K., Nguyen L.T., Van Nguyen L., Nguyen T.N., Nguyen T.T., Van Pham D., Nguyen D.H., Nguyen T.D., To T.L., Van Nguyen H., Kida H., Sakoda Y. Genetic and antigenic characterization of H5, H6 and H9 avian influenza viruses circulating in live bird markets with intervention in the center part of Vietnam. Vet. Microbiol. 2016;192:194–203. doi: 10.1016/j.vetmic.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Conenello G.M., Zamarin D., Perrone L.A., Tumpey T., Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin E.R., Friedman K., Krauss S., Nolting J.M., Poulson R.L., Slemons R.D., Stallknecht D.E., Webster R.G., Bowman A.S. Subtype diversity of influenza A virus in North American Waterfowl: a multidecade study. J Virol. 2020;94:e02022–02019. doi: 10.1128/JVI.02022-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis R.O., Bean W.J., Kawaoka Y., Webster R.G. Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology. 1989;169:408–417. doi: 10.1016/0042-6822(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Fourment M., Holmes E.C. Avian influenza virus exhibits distinct evolutionary dynamics in wild birds and poultry. BMC Evol Biol. 2015;15:120. doi: 10.1186/s12862-015-0410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gischke M., Ulrich R., I. F. O. Scheibner D., Salaheldin A.H., Crossley B., Bottcher-Friebertshauser E., Veits J., Mettenleiter T.C., Abdelwhab E.M. Insertion of basic amino acids in the hemagglutinin cleavage site of H4N2 Avian Influenza Virus (AIV)-reduced virus fitness in chickens is restored by reassortment with highly pathogenic H5N1 AIV. Int J Mol Sci. 2020;21:2353. doi: 10.3390/ijms21072353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Isoda N., Bazarragchaa E., Nomura N., Matsuno K., Okamatsu M., Kida H., Sakoda Y. Potency of an inactivated influenza vaccine against a challenge with A/Swine/Missouri/A01727926/2015 (H4N6) in mice for pandemic preparedness. Vaccines. 2020;8 doi: 10.3390/vaccines8040768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander L.P., Fojtik A., Kienzle-Dean C., Davis-Fields N., Poulson R.L., Davis B., Mowry C., Stallknecht D.E. Prevalence of influenza A viruses in ducks sampled in northwestern minnesota and evidence for predominance of H3N8 and H4N6 subtypes in Mallards, 2007-2016. Avian Dis. 2019;63:126–130. doi: 10.1637/11851-041918-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Liu X., Li S., Guo X., Yang Y., Jin M. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. J Virol. 2012;86:13879. doi: 10.1128/JVI.02726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Couceiro J.N., Kelm S., Baum L.G., Krauss S., Castrucci M.R., Donatelli I., Kida H., Paulson J.C., Webster R.G., Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P., Wei L., Song Y., Cui J., Song H., Cao L., Yuan R., Luo K., Liao M. D701N mutation in the PB2 protein contributes to the pathogenicity of H5N1 avian influenza viruses but not transmissibility in guinea pigs. Front. Microbiol. 2014;5:642. doi: 10.3389/fmicb.2014.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal R.P., Alymova I.V., York I.A. Evolution and virulence of influenza A Virus protein PB1-F2. Int. J. Mol. Sci. 2017;19:96. doi: 10.3390/ijms19010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Choi J.G., Kim K.I., Park H.Y., Park C.K., Lee Y.J. Genetic and antigenic characteristics of H4 subtype avian influenza viruses in Korea and their pathogenicity in quails, domestic ducks and mice. J. Gen. Virol. 2013;94:30–39. doi: 10.1099/vir.0.046581-0. [DOI] [PubMed] [Google Scholar]
- Karasin A.I., Brown I.H., Carman S., Olsen C.W. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J. Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasin A.I., Olsen C.W., Brown I.H., Carman S., Stalker M., Josephson G. H4N6 influenza virus isolated from pigs in Ontario. Can. Vet. J. 2000;41:938–939. [PMC free article] [PubMed] [Google Scholar]
- Kayali G., Barbour E., Dbaibo G., Tabet C., Saade M., Shaib H.A., Debeauchamp J., Webby R.J. Evidence of infection with H4 and H11 avian influenza viruses among Lebanese chicken growers. PloS one. 2011;6:e26818. doi: 10.1371/journal.pone.0026818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayali G., Ortiz E.J., Chorazy M.L., Gray G.C. Evidence of previous Avian influenza infection among US turkey workers. Zoonoses Public Health. 2009;57:265–272. doi: 10.1111/j.1863-2378.2009.01231.x. [DOI] [PubMed] [Google Scholar]
- Kayali G., Ortiz E.J., Chorazy M.L., Gray G.C. Evidence of previous avian influenza infection among US turkey workers. Zoonoses Public Health. 2010;57:265–272. doi: 10.1111/j.1863-2378.2009.01231.x. [DOI] [PubMed] [Google Scholar]
- Khan S.U., Gurley E.S., Gerloff N., Rahman M.Z., Simpson N., Rahman M., Haider N., Chowdhury S., Balish A., Zaman R.U., Nasreen S., Das B.C., Azziz-Baumgartner E., Sturm-Ramirez K., Davis C.T., Donis R.O., Luby S.P. Avian influenza surveillance in domestic waterfowl and environment of live bird markets in Bangladesh, 2007-2012. Sci. Rep. 2018;8:9396. doi: 10.1038/s41598-018-27515-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Margalef N., Tolf C., Grosbois V., Avril A., Bengtsson D., Wille M., Osterhaus A.D.M.E., Fouchier R.A.M., Olsen B., Waldenström J. Long-term variation in influenza A virus prevalence and subtype diversity in migratory mallards in northern Europe. Proc. Royal Society B. 2014;281 doi: 10.1098/rspb.2014.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.K., Kang H.M., Song B.M., Lee Y.N., Heo G.B., Lee H.S., Lee Y.J., Kim J.H. Surveillance of avian influenza viruses in South Korea between 2012 and 2014. Virol. J. 2017;14:54. doi: 10.1186/s12985-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Deng G., Shi J., Wang S., Zhang Q., Kong H., Gu C., Guan Y., Suzuki Y., Li Y., Jiang Y., Tian G., Liu L., Li C., Chen H., Dermody T.S. Genetics, receptor binding, replication, and mammalian transmission of H4 Avian Influenza viruses isolated from live poultry markets in China. J. Virol. 2016;90:1455–1469. doi: 10.1128/JVI.02692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., He S., Walker D., Zhou N., Perez D.R., Mo B., Li F., Huang X., Webster R.G., Webby R.J. The influenza virus gene pool in a poultry market in South central china. Virology. 2003;305:267–275. doi: 10.1006/viro.2002.1762. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhu W., Feng Z., Gao R., Guo J., Li X., Liu J., Wang D., Shu Y. Substitution of D701N in the PB2 protein could enhance the viral replication and pathogenicity of Eurasian avian-like H1N1 swine influenza viruses. Emerg. Microbes Infect. 2018;7:75. doi: 10.1038/s41426-018-0073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhuang Q., Wang S., Jiang W., Jin J., Peng C., Hou G., Li J., Yu J., Yu X., Liu H., Sun S., Yuan L., Chen J. Control of avian influenza in China: Strategies and lessons. Transbound. Emerg. Dis. 2020;67:1463–1471. doi: 10.1111/tbed.13515. [DOI] [PubMed] [Google Scholar]
- Long J.S., Mistry B., Haslam S.M., Barclay W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019;17:67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- Lublin A., Thie N., Shkoda I., Simanov L., Bar-Gal G.K., Farnoushi Y., King R., Getz W.M., Kamath P.L., Bowie R.C.K., Nathan R. First detection of avian influenza subtype H4N6 in Israel in a wild mallard (Anas platyrhynchos) Transbound. Emerg. Dis. 2022;69:e3316–e3326. doi: 10.1111/tbed.14610. [DOI] [PubMed] [Google Scholar]
- Luo M. Influenza virus entry. Adv. Exp. Med. Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Xie Z., Li M., Li D., Xie L., Huang J., Zhang M., Zeng T., Wang S., Fan Q., Zhang Y., Xie Z., Deng X., Liu J. Survey of low pathogenic avian influenza viruses in live poultry markets in Guangxi Province, Southern China, 2016-2019. Sci. Rep. 2021;11:23223. doi: 10.1038/s41598-021-02639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y., Kida H., Yanagowa R. Isolation of an influenza virus subtype Hav4Navl from a budgerigar. Microbiol. Immunol. 1979;23:35–38. doi: 10.1111/j.1348-0421.1979.tb00438.x. [DOI] [PubMed] [Google Scholar]
- Morales A.C., Jr, Hilt D.A., Williams S.M., Pantin-Jackwood M.J., Suarez D.L., Spackman E., Stallknecht D.E., Jackwood M.W. Biologic characterization of H4, H6, and H9 type low pathogenicity avian influenza viruses from wild birds in chickens and turkeys. Avian Dis. 2009;53:552–562. doi: 10.1637/8877-041509-Reg.1. [DOI] [PubMed] [Google Scholar]
- Ninomiya A., Takada A., Okazaki K., Shortridge K.F., Kida H. Seroepidemiological evidence of avian H4, H5, and H9 influenza A virus transmission to pigs in southeastern China. Vet. Microbiol. 2002;88:107–114. doi: 10.1016/s0378-1135(02)00105-0. [DOI] [PubMed] [Google Scholar]
- Olsen B., Munster V.J., Wallensten A., Waldenström J., Osterhaus A.D., Fouchier R.A. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Ornelas-Eusebio E., Obregon-Ascencio A., Chavez-Maya F., Garcia-Espinosa G. Molecular characterization of an influenza A virus (H4N2) isolated from waterfowl habitats in the State of Mexico. J. Vet. Med. Sci. 2015;77:365–369. doi: 10.1292/jvms.14-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar S.D., Kale S.D., Rawankar A.S., Koratkar S.S., Raut C.G., Pande S.A., Mullick J., Mishra A.C. Avian influenza surveillance reveals presence of low pathogenic avian influenza viruses in poultry during 2009-2011 in the West Bengal State, India. Virol. J. 2012;9:151. doi: 10.1186/1743-422X-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar S.D., Keng S.S., Kode S.S., Tare D.S., Singh D.K., Mullick J. A novel reassortant avian influenza H4N6 virus isolated from an environmental sample during a surveillance in Maharashtra, India. Indian J Med Res. 2021;154:871–887. doi: 10.4103/ijmr.IJMR_1514_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralow A., Hoffmann M., Nguyen-Khuong T., Pioch M., Hennig R., Genzel Y., Rapp E., Reichl U. Comprehensive N-glycosylation analysis of the influenza A virus proteins HA and NA from adherent and suspension MDCK cells. FEBS J. 2021;288:4869–4891. doi: 10.1111/febs.15787. [DOI] [PubMed] [Google Scholar]
- Quan C., Huang T., Chen X., Zhang J., Wang Q., Zhang C., Zhang T., Zhou L., Shu L., Long C., Yang L., Du X., Zhao Y., Liu P., Song H., Shi W., Bi Y., Lv Q., Liu W.J., Gao G.F. Genomic characterizations of H4 subtype avian influenza viruses from live poultry markets in Sichuan province of China, 2014-2015. Sci. China Life Sci. 2018;61:1123–1126. doi: 10.1007/s11427-018-9327-4. [DOI] [PubMed] [Google Scholar]
- Reid S.M., Brookes S.M., Nunez A., Banks J., Parker C.D., Ceeraz V., Russell C., Seekings A., Thomas S.S., Puranik A., Brown I.H. Detection of non-notifiable H4N6 avian influenza virus in poultry in Great Britain. Vet. Microbiol. 2018;224:107–115. doi: 10.1016/j.vetmic.2018.08.026. [DOI] [PubMed] [Google Scholar]
- Shi Y., Cui H., Wang J., Chi Q., Li X., Teng Q., Chen H., Yang J., Liu Q., Li Z. Characterizations of H4 avian influenza viruses isolated from ducks in live poultry markets and farm in Shanghai. Sci. Rep. 2016;6:37843. doi: 10.1038/srep37843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y.J., Lee I.W., Kim E.H., Kim Y.I., Kwon H.I., Park S.J., Nguyen H.D., Kim S.M., Kwon J.J., Choi W.S., Beak Y.H., Song M.S., Kim C.J., Webby R.J., Choi Y.K. Genetic characterisation of novel, highly pathogenic avian influenza (HPAI) H5N6 viruses isolated in birds, South Korea, November 2016. Euro surveillance. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.1.30434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton R.M., Huber V.C. Comparing influenza virus biology for understanding influenza D virus. Viruses. 2022;14:1036. doi: 10.3390/v14051036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Qi J., Xiao H., Bi Y., Zhang W., Xu Y., Wang F., Shi Y., Gao G.F. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep. 2017;20:1201–1214. doi: 10.1016/j.celrep.2017.07.028. [DOI] [PubMed] [Google Scholar]
- Song W., Wang P., Mok B.W., Lau S.Y., Huang X., Wu W.L., Zheng M., Wen X., Yang S., Chen Y., Li L., Yuen K.Y., Chen H. The K526R substitution in viral protein PB2 enhances the effects of E627K on influenza virus replication. Nat Commun. 2014;5:5509. doi: 10.1038/ncomms6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallknecht D.E., Fojtik A., Carter D.L., Crum-Bradley J.A., Perez D.R., Poulson R.L. Naturally acquired antibodies to influenza A virus in fall-migrating North American mallards. Vet Sci. 2022;9:214. doi: 10.3390/vetsci9050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Qi W.B., Chen J.D., Cao N., Zhu W.J., Yuan L.G., Wang H., Zhang G.H. Complete genome sequence of an avian-like H4N8 swine influenza virus discovered in southern China. J Virol. 2012;86:9542. doi: 10.1128/JVI.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Świętoń E., Tarasiuk K., Śmietanka K. Low pathogenic avian influenza virus isolates with different levels of defective genome segments vary in pathogenicity and transmission efficiency. Vet. Res. 2020;51:108. doi: 10.1186/s13567-020-00833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veits J., Weber S., Stech O., Breithaupt A., Gräber M., Gohrbandt S., Bogs J., Hundt J., Teifke J.P., Mettenleiter T.C., Stech J. Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. Proc. Natl Acad Sci. USA. 2012;109:2579–2584. doi: 10.1073/pnas.1109397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Li M., Xiong C., Yan Y., Hu J., Hao M., Liang B., Chen J., Chen G., Yang G., Li Y., Zhang J., Gulyaeva M., Shestopalov A., Shi W., Bi Y., Liu H., Wang H., Liu D., Chen J. Ecology of avian influenza viruses in migratory birds wintering within the Yangtze River wetlands. Sci. Bull (Beijing) 2021;66:2014–2024. doi: 10.1016/j.scib.2021.03.023. [DOI] [PubMed] [Google Scholar]
- Wille M., Brojer C., Lundkvist A., Jarhult J.D. Alternate routes of influenza A virus infection in Mallard (Anas platyrhynchos) Vet. Res. 2018;49:110. doi: 10.1186/s13567-018-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M., Tolf C., Latorre-Margalef N., Fouchier R.A.M., Halpin R.A., Wentworth D.E., Ragwani J., Pybus O.G., Olsen B., Waldenström J. Evolutionary features of a prolific subtype of avian influenza A virus in European waterfowl. Virus Evol. 2022;8:veac074. doi: 10.1093/ve/veac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.S., Yoon S.W., Zanin M., Song M.S., Oshansky C., Zaraket H., Sonnberg S., Rubrum A., Seiler P., Ferguson A., Krauss S., Cardona C., Webby R.J., Crossley B. Characterization of an H4N2 influenza virus from Quails with a multibasic motif in the hemagglutinin cleavage site. Virology. 2014;468:72–80. doi: 10.1016/j.virol.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Peng X., Peng X., Cheng L., Lu X., Jin C., Xie T., Yao H., Wu N. Genetic characterization of natural reassortant H4 subtype avian influenza viruses isolated from domestic ducks in Zhejiang province in China from 2013 to 2014. Virus Genes. 2015;51:347–355. doi: 10.1007/s11262-015-1245-2. [DOI] [PubMed] [Google Scholar]
- Xu G., Wang F., Li Q., Bing G., Xie S., Sun S., Bian Z., Sun H., Feng Y., Peng X., Jiang H., Zhu L., Fan X., Qin Y., Ding J. Mutations in PB2 and HA enhanced pathogenicity of H4N6 avian influenza virus in mice. J. Gen. Virol. 2020;101:910–920. doi: 10.1099/jgv.0.001192. [DOI] [PubMed] [Google Scholar]
- Xu Y., Tang L., Gu X., Bo S., Ming L., Ma M., Zhao C., Sun K., Liu Y., He G. Characterization of avian influenza A (H4N2) viruses isolated from wild birds in Shanghai during 2019 to 2021. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2023.102948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Zheng H., Xiong J., Ma L., Gui R., Zhu G., Li Y., Yang G., Chen G., Zhang J., Chen Q. Genetic and pathogenic characterization of Avian Influenza virus in migratory birds between 2015 and 2019 in Central China. Microbiol Spectr. 2022;10 doi: 10.1128/spectrum.01652-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.Y., Wang Y.L., Yu K.X., Zhang Y.X., Xu H.Y., Yang J.X., Li F., Song M.X. Isolation and genetic characterization of avian influenza virus H4N6 from ducks in China. Arch Virol. 2015;160:55–59. doi: 10.1007/s00705-014-2229-6. [DOI] [PubMed] [Google Scholar]
- Zamarin D., Ortigoza M.B., Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol. 2006;80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chen Q., Chen Z. Characterization of an H4N2 avian influenza virus isolated from domestic duck in Dongting Lake wetland in 2009. Virus Genes. 2012;44:24–31. doi: 10.1007/s11262-011-0658-9. [DOI] [PubMed] [Google Scholar]