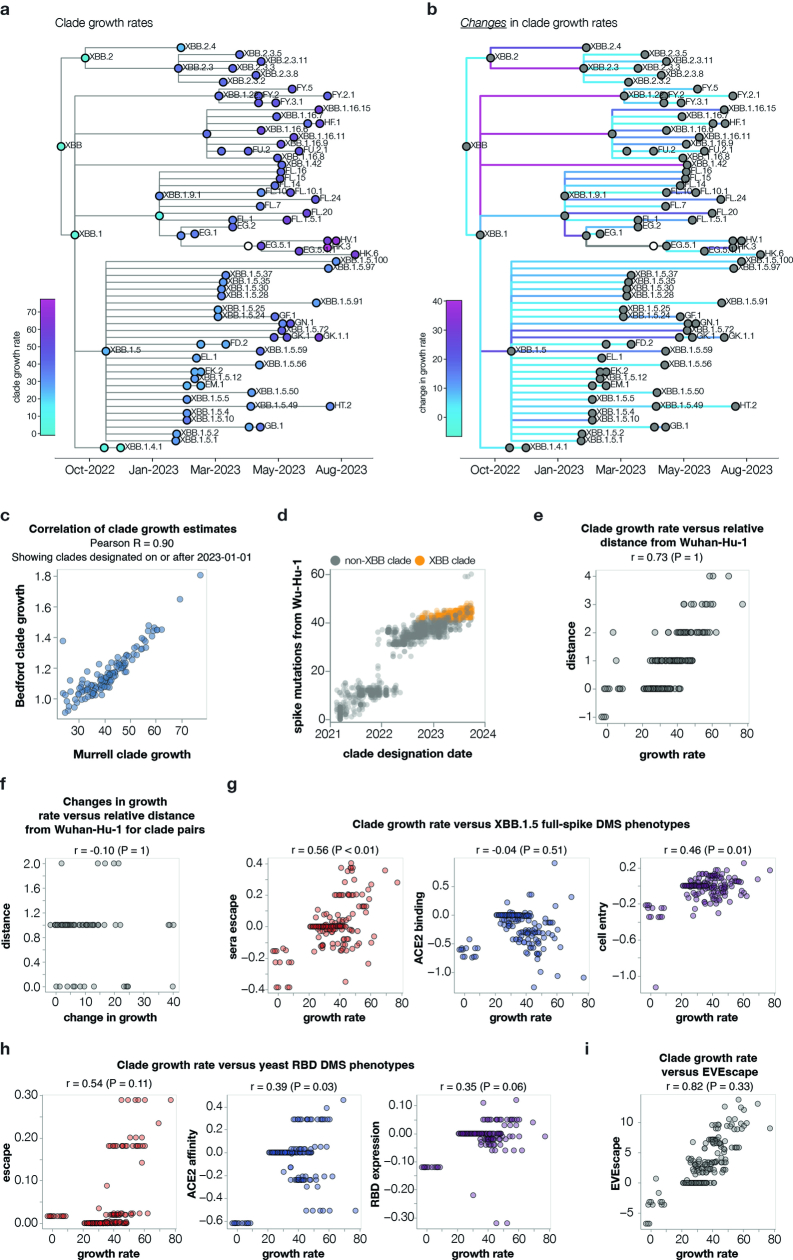
Extended Data Fig. 10. Correlations in absolute clade growth with absolute clade phenotypes.
a, Phylogenetic tree of XBB-descended Pango clades, colored by their relative growth rates. The tree shows only clades with at least 400 sequences and at least one new spike mutation, and their ancestors. Ancestor clades with insufficient sequences for growth rate estimates are in white. b, The same phylogeny but with branches colored by the change in growth rate between parent-descendant clade pairs. c, Correlation between clade growth estimates made using the Murrell lab multinomial logistic regression model (see methods) or a hierarchical multinomial logistic regression implemented by the Bedford lab68 (see https://github.com/nextstrain/forecasts-ncov/). Both sets of estimates are for clades designated after Jan-1-2023 and use the data available as of Oct-2-2023. The estimates are highly correlated, and everywhere else in this paper we report analyses using the Murrell lab estimates. d, Number of spike amino-acid mutations relative to the early Wuhan-Hu-1 virus in all SARS-CoV-2 Pango clades versus the clade designation dates. XBB-descended clades are in orange. As can be seen from this plot, newer clades tend to have more spike mutations. e, Because newer clades tend to have both more mutations and better growth, clade growth rate is trivially correlated with a clade’s relative distance (number of spike mutations) from Wuhan-Hu-1. However, this correlation is not informative as it is already known that new clades tend to have more mutations. f, If we instead correlate the change in growth rate between parent-descendant clade pairs separated by at least one spike mutation (Fig. 6b) with the change in spike mutational distance to Wuhan-Hu-1 there is no correlation, since this approach removes the co-variation with total mutation count. Therefore, simple mutation counting is not informative for predicting changes in clade growth. g, Correlations for the phenotypes measured by the full spike deep mutational scanning in the current paper; h, the phenotypes measured in yeast display RBD deep mutational scanning; i, predicted by the EVEscape method. These plots differ from Fig. 6a and Extended Data Fig. 11 in that they show the correlations in absolute clade growth with the absolute clade phenotypes, rather than comparing the changes in both for each parent-descendant clade pair. Absolute clade phenotypes are computed as the sum of mutation effects. The P-values above the plots is a one sided test that computes the fraction of times the correlation is greater than that for the actual data after randomizing the phenotypic effects among mutations. Note that the correlations are not reflective of the P-values (there can be high correlations but non-significant P-values) for the reasons noted in the main text and in e—phylogenetic correlations, and the fact that new clades have both more mutations and higher growth so that any “phenotype” that amounts to counting mutations gives a correlation in these plots. For this reason, comparing changes in clade growth to changes in spike phenotypes as done in Fig. 6a and Extended Data Fig. 11 is the correct approach to test whether a method can actually predict which new clades will be successful.