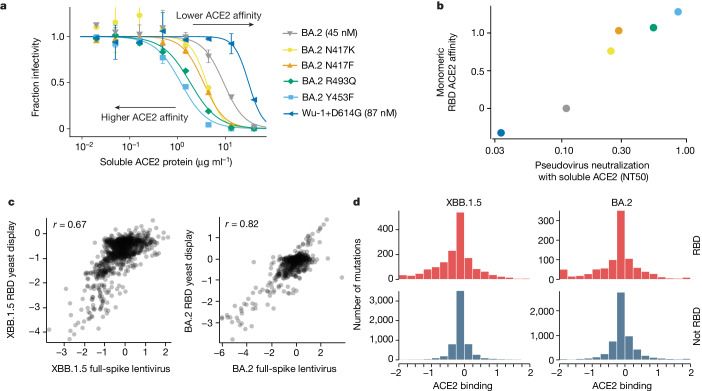
Fig. 2. Effects of mutations on full-spike ACE2 binding measured using pseudovirus deep mutational scanning.
a, Neutralization of pseudoviruses with the indicated spikes by soluble monomeric ACE2. Viruses with spikes that have stronger binding to ACE2 are neutralized more efficiently by soluble ACE2 (lower half-maximal neutralizing titers(NT50)), whereas viruses with spikes with worse binding are neutralized more weakly. Error bars indicate standard error between two replicates. ACE2 affinity values measured by surface plasmon resonance for BA.2 and Wu-1+D614G are shown in brackets18. b, Correlation between neutralization NT50 by soluble ACE2 versus the RBD affinity for ACE2 as measured by titrations using yeast-displayed RBD2. c, Correlations between the effects of RBD mutations on ACE2 binding measured using the pseudovirus-based approach (this study) and yeast-based RBD display2,20. d, Distribution of effects of individual mutations on full-spike ACE2 binding for all functionally tolerated mutations in our libraries, stratified by RBD versus non-RBD mutations. Note that effects of magnitude greater than two are clamped to the limits of the plots’ x axes. The effects of individual XBB.1.5 spike mutations on ACE2 binding are shown in Extended Data Fig. 3.