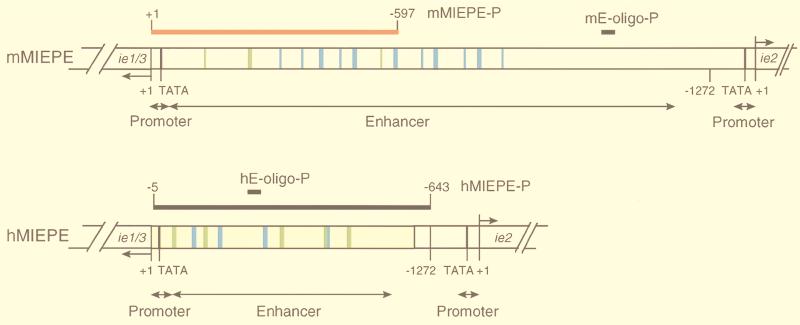
FIG. 3.
Map of MIEPE type-specific hybridization probes. (Top) Structural organization of the authentic MIEPE of mCMV (mMIEPE), essentially based on data by Dorsch-Häsler et al. (11). Map positions refer to the 5′ start site (counted as +1) of the ie1/3 transcription unit. The map location of oligonucleotide probe mE-oligo-P used for Southern blot hybridization (see Fig. 1B) is indicated. The red bar represents the polynucleotide probe mMIEPE-P used for mCMV MIEPE-specific red staining in the two-color ISH. (Bottom) Structural organization of the chimeric region in recombinant virus mCMVhMIEPE. Sequences of the inserted hCMV core promoter and enhancer (hMIEPE), representing positions −14 to −601 in hCMV, are highlighted by yellow shading. Map positions refer to the 5′ start site (counted as +1) of the mCMV ie1/3 transcription unit. The map location of oligonucleotide probe hE-oligo-P (see Fig. 1B) is indicated. The long black bar represents the polynucleotide probe hMIEPE-P used for hCMV MIEPE-specific black staining in the two-color ISH. The two maps are drawn to scale. Consensus sequences within 18- and 19-bp direct repeats, encompassing NF-κB and CREB/ATF binding sites, are marked by blue and green coloring, respectively.