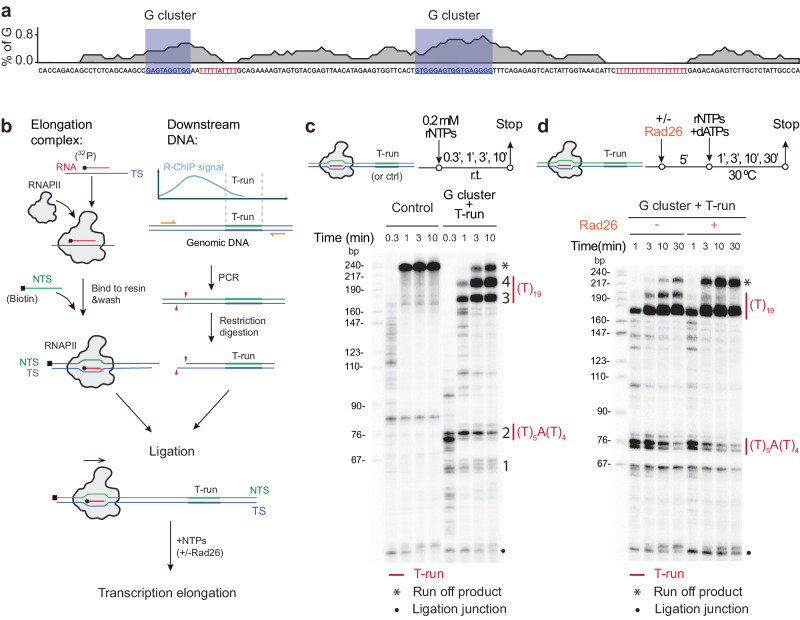
Fig. 3. CSB resolves RNAPII Pausing at T-runs.
a Sequence feature of the DNA template used for the run-off assay. Two G clusters are shaded in blue box and Two T-runs are highlighted in red. b A scheme of the experimental setup to study the function of CSB in RNAPII transcription in vitro. An RNAPII elongation complex (EC) segment and a T-run containing DNA fragment (based on the R-loop identified from R-ChIP) were reconstituted individually and ligated by T4 ligase. A biotin label is shown as a black square. c RNAPII pausing at T-run. The top section displays a schematic of the transcription reaction. Below shows the run-off signals, illustrating multiple pausing sites during RNAPII transcription on a control sequence lacking T-runs (Left) compared to a template with T-run associated R-loops (Right). The pausing sites and T-run regions are indicated on the right. Ligation truncation corresponds to transcripts from reconstituted ECs that failed to ligate with the downstream T-run associated R-loop template. d Role of Rad26 in promoting RNAPII bypassing the T-run. The top section depicts a schematic of the transcription reaction. Below shows specific RNAPII pausing sites during RNAPII elongation along a T-run associated R-loop template without (left) or with (right) purified Rad26. Experiments in c, d were repeated independently three times with similar results.