Abstract
Background
The SPF10 LiPA-25 system for human papillomavirus (HPV) detection with high analytical performance is widely used in HPV vaccine clinical trials. To develop and evaluate more valent HPV vaccines, other comparable methods with simpler operations are needed.
Methods
The performance of the LiPA-25 against that of other 7 assays, including 4 systems based on reverse hybridization (Bohui-24, Yaneng-23, Tellgen-27, and Hybribio-16) and 3 real-time polymerase chain reaction (PCR) assays (Hybribio-23, Bioperfectus-21, and Sansure-26), was evaluated in selected 1726 cervical swab and 56 biopsy samples. A total of 15 HPV genotypes (HPV 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66) were considered for comparison for each HPV type.
Results
Among the swab samples, compared to LiPA-25, compatible genotypes were observed in 94.1% of samples for Hybribio-23, 92.8% for Yaneng-23, 92.6% for Bioperfectus-21, 92.4% for Hybribio-16, 91.3% for Sansure-26, 89.7% for Bohui-24, and 88.0% for Tellgen-27. The highest overall agreement of the 15 HPV genotypes combined was noted for Hybribio-23 (κ = 0.879, McNemar's test: P = 0.136), followed closely by Hybribio-16 (κ = 0.877, P< 0.001), Yaneng-23 (κ = 0.871, P < 0.001), Bioperfectus-21 (κ = 0.848, P < 0.001), Bohui-24 (κ = 0.847, P < 0.001), Tellgen-27 (κ = 0.831, P < 0.001), and Sansure-26 (κ = 0.826, P < 0.001). Additionally, these systems were also highly consistent with LiPA-25 for biopsy specimens (all, κ > 0.897).
Conclusions
The levels of agreement for the detection of 15 HPV types between other 7 assays and LiPA-25 were all good, and Hybribio-23 was most comparable to LiPA-25. The testing operation of HPV genotyping should also be considered for vaccine and epidemiological studies.
Keywords: Human papillomavirus, Vaccine, Cervical cancer, Line probe assay, HPV genotyping, Polymerase chain reaction
1. Introduction
The incidence rate of cervical cancer has decreased in many countries for decades, while it has increased in China.1 In 2018, World Health Organization (WHO) called for action toward the elimination of cervical cancer. Human papillomavirus (HPV) vaccine is one of two powerful weapons against HPV infection.2 HPV infection is associated with benign and malignant lesions of the cutaneous and mucosal epithelia. Persistent infection with high-risk HPV (HR-HPV) genotypes is the leading cause of the development of cervical cancer.3,4 Epidemiologic research studies have classified 14 HPV genotypes as high risk, based on their association with cervical cancer, i.e.,16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68.5,6 Among them, two HR-HPVs (HPV 16 and 18) are the causative factors for about 70% of cervical cancers. Another two low-risk (LR) HPV genotypes (HPV 6 and 11) cause 90% of genital warts, and most of them require treatment.7 HPV vaccination and screening programs are effective strategies in disease prevention and elimination.2,8
To evaluate the effects of type-specific preventive or therapeutic vaccination in the population, sensitive and specific HPV genotyping methods are critical for the selection and monitoring of study subjects. Although several HPV detection methods could monitor the oncogenic vaccine types,9, 10, 11 the original SPF10 Line Probe Assay (LiPA-25) system has good analytical sensitivity and specificity for HPV genotyping in clinical specimens12,13 and was widely applied in HPV vaccine as a golden standard for determining bivalent vaccine efficacy14, 15, 16, 17, 18, 19, 20 and epidemiologic HPV studies in the world.21,22 The system is based on broad-spectrum polymerase chain reaction (PCR) assay for the amplification of a 65-bp fragment in the L1 region, and a reverse hybridization assay, which allows the detection of 13 individual HR-HPV genotypes (i.e., HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66), 11 individual LR-HPV genotypes (i.e., HPV 6, 11, 34, 40, 42, 43, 44, 53, 54, 70, and 74), and a compound genotype (i.e., HPV 68/73).23
In the last several years, advances have been made in HPV detection methods, and various PCR-based genotyping methods with primers aimed at the late region (L1 or L2) or the early region (E1, E2, E4, E6, or E7) of the viral genome have been reported.24, 25, 26 These products mainly consist of real-time fluorescence PCR assays with type-specific primers,24, 25, 26, 27 or using tagging oligonucleotide cleavage and extension (TOCE) technology,28,29 and the general amplification reactions with consensus PCR primer sets that subsequently are detected by type-specific probe hybridization24,30, 31, 32 or sequencing.33,34 However, limited studies with representative samples were conducted to evaluate the performance of these methods.
We performed a blind and head-to-head study by using selected cervical samples, with a focus on HPV types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66, to compare seven multiplex HPV tests with SPF10 LiPA-25 for finding the eligible method to apply for HPV vaccine efficacy and epidemiologic studies.
2. Materials and methods
2.1. Subjects
Cervical swab and formalin-fixed, paraffin-embedded (FFPE) biopsy specimens were provided by the Department of Cancer Epidemiology in Cancer Hospital, Chinese Academy of Medical Sciences. A total of 1,897 cervical swab specimens were collected in Thinprep® Pap Test PreservCyt® solution and stored at -80°C, of which the HPV-status was determined with the SPF10 LiPA-25, were selected and prepared 500 μl aliquots for 7 other assays. In addtion, we colleced 56 FFPE samples diagnosed with CIN+, including cervical intraepithelial neoplasia grade 3 (CIN3; n = 3), cervical micro-invasive carcinoma (n = 2), squamous-cell carcinoma (SCC; n = 49), and adenocarcinoma (n = 2). To confirm the presence of CIN or worse in the FFPE samples used for HPV DNA analysis, a sandwich sectioning method was used.35 The outer sections were stained with hematoxylin and eosin for histological diagnosis, while the inner sections were used for HPV detection by the 8 systems. Seven systems of HPV DNA detection were selected for this study and named with “company name” plus “the number of detected HPV genotypes,” i.e., Bohui-24, Yaneng-23, Tellgen-27, Hybribio-16, Hybribio-23, Bioperfectus-21, and Sansure-26.
2.2. Plasmids
To compare the sensitivities of these assays in the limit of detection analysis, a 10-fold dilution series of the HPV L1, HPV E6, and HPV E7 plasmids purified in a background of human genomic DNA (human placenta, SIGMA-ALDRICH®) for 15 HPV genotypes (6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66), were tested in 8 assays. The original concentration of HPV DNA was 107 copies/ml. These plasmids were provided by the National Institutes for Food and Drug Control of China. The limit of detection for 15 HPV genotypes represented either 10 to 1000 copies/test for LiPA-25, 60 to 600 copies/test for Bohui-24, 5 to 500 copies/test for Yaneng-23, 20 to 500 copies/test for Tellgen-27, 10 to 1000 copies/test for Hybribio-16, 20 to 2000 copies/test for Hybribio-23, 20 to 2000 copies/test for Bioperfectus-21, and 5 to 500 copies/test for Sansure-26 (Supplementary Table 1).
2.3. HPV genotyping tests
These methods have different procedures for DNA extraction, PCR, and genotyping test, which were performed according to the manufacturer's instructions. Detailed methods are provided in the Supplementary Materials. In this study, the 8 genotyping systems were divided into two methods: the hybridization method and the real-time fluorescence PCR method.
2.3.1. Hybridization methods by LiPA-25, Bohui-24, Yaneng-23, Hybribio-16, and Tellgen-27
Magnetic bead-based DNA extraction for swab samples or DNA extraction by the boiling method for FFPE samples was performed. Broad-spectrum primers target on L1 region were applied for PCR amplification, and amplicons were genotyped by reverse hybridization with linear/two-dimension probe arrays on membrane of LiPA-25, Bohui-24, Yaneng-23, and Hybribio-16, or with multiplex Luminex bead-based probe array of Tellgen-27 (Table 1 and Supplementary Table 2).
Table 1.
Comparison of the main characteristics of 8 HPV genotyping systems.
| System |
Methodology of DNA extraction |
PCR and HPV genotyping method |
Internal control (human DNA) | ||||
|---|---|---|---|---|---|---|---|
| Swab | Biopsy | Primer | Targeted region | Amplimer length (bp) | Detection method | ||
| LiPA-25 | Magnetic bead-based | Boiling | Broad-spectrum | L1: SPF10 | ∼ 65 | Reverse hybridization | No |
| Bohui-24 | Magnetic bead-based | Boiling | Broad-spectrum | L1: 6280∼6540 | ∼ 260 | Reverse hybridization | Yes |
| Yaneng-23 | Magnetic bead-based | Boiling | Broad-spectrum | L1: GP5+/6+ | ∼ 135 | Reverse hybridization | Yes |
| Hybribio-16 | Magnetic bead-based | Boiling | Broad-spectrum | L1: MY09/11 | ∼ 450 | Flow cytometry hybridization | Yes |
| Tellgen-27 | Chelex®100-based | Boiling + Chelex®100-based | Broad-spectrum | L1: MY09/11 | ∼ 450 | Real-time PCR | Yes |
| Hybribio-23 | Magnetic bead-based | Boiling | Multiplex type-specific | L1/L2/E1/E2/E4/E6/E7 | ∼ 150 | Real-time PCR | Yes |
| Bioperfectus-21 | Magnetic bead-based | Boiling + Magnetic bead-based | Multiplex type-specific | L1/E1/E2/E7 | ∼ 120 | Real-time PCR | Yes |
| Sansure-26 | One-step | Boiling + Magnetic bead-based | Multiplex type-specific | L1/L2/E1/E6/E7 | ∼ 200 | Real-time PCR | Yes |
Hybribio-23: HPV types, CT≤40; Internal control, CT≤40; Positive control, CT≤36; Negative control, undetected.
Bioperfectus-21: HPV types (HPV6, CT<37.4; HPV11, CT<37.9; HPV16, CT<38.0; HPV18, CT<37.8; HPV31, CT<36.2; HPV33, CT<37.2; HPV35, CT<37.2; HPV39, CT<37.6; HPV45, CT<36.4; HPV51, CT<38.1; HPV52, CT<37.8; HPV56, CT<37.7; HPV58, CT<37.9; HPV59, CT<37.5; HPV66, CT<38.0); Internal control, CT≤36.7; Positive control, CT≤30; Negative control, undetected.
Sansure-26: HPV types, CT≤39; Internal control, CT≤40; Positive control; CT≤36, Negative control, undetected.
Abbreviation: HPV, human papillomavirus; PCR, polymerase chain reaction.
2.3.2. Real-time PCR detection by Hybribio-23, Bioperfectus-21, and Sansure-26
For Hybribio-23 and Bioperfectus-21, magnetic bead-based DNA extraction was used for swab samples, which is different from that of Sansure-26’s one-step technology with lysis buffer for the direct and rapid release of DNA. Boiling method or magnetic bead-based DNA extraction was used for FFPE samples. Real-time PCR with type-specific primers targeted on L1/L2/E1/E2/E4/E6/E7 was used for HPV genotyping (Table 1 and Supplementary Table 2).
2.4. Statistical analysis
Only the 15 HPV genotypes (i.e., HPV types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66) jointly detected by 8 methods were included for analysis. LiPA-25 was used as a reference for comparison with the other 7 methods. Chi-square test was used to evaluate the difference between LiPA25 and other 7 assays by infection status. When the specimens were analyzed by the two methods (LiPA-25 and other system), genotyping results of specimens were categorized into 3 groups: concordant (100% identical), compatible (at least one genotype of multiple infection was detected by both assays), and discordant.36 Regarding HPV types, the proportion of positive agreement (Ppos) and the proportion of negative agreement (Pneg) were calculated as reported previously,25 and two-tailed McNemar's test was used for mutual comparisons. The level of agreement was determined using Cohen's kappa statistics. The level of statistical significance was set at 0.05. All analyses were performed using R language software (R version 3.5.3).
3. Results
3.1. Genotyping agreement on cervical swab and biopsy samples
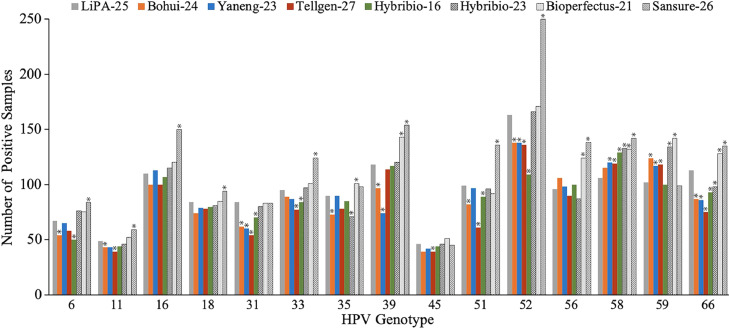
The data of 15 genotypes jointly detected by 8 systems were used for analysis. For swab samples, 1,897 women were selected for this study, 171 samples were not included for analysis due to the defects of samples, and finally, 1,726 samples were included for data analysis. The number of invalid detections was 0 for LiPA-25, 1 for Bohui-24, 7 for Yaneng-23, 27 for Tellgen-27, 15 for Hybribio-16, 11 for Hybribio-23, 0 for Bioperfectus-21, and 0 for Sansure-26, and HPV DNA positivity was 58.0% (1,001/1,726), 50.9% (878/1,725), 54.0% (929/1,719), 50.1% (851/1,687), 53.3% (911/1,710), 56.4% (967/1,715), 58.9% (1,016/1,726), and 62.9% (1,086/1,726), correspondingly (Fig. 1 and Table 2).
Fig. 1.
Positives by HPV genotype from LiPA-25 and other 7 assays for cervical swab specimens. Significant differences (P < 0.05, McNemar's test) of other assays (Sansure-26, Bioperfectus-21, Bohui-24, Yaneng-23, Tellgen-27, Hybribio-23, and Hybribio-16) and LiPA-25 are indicated by asterisks.
Table 2.
Distribution of the number of 15 HPV genotypesa detected by cervical swab and biopsy specimens using LiPA-25 and other 7 assays.
| Methods | No. of samples | 0 type (%) | All positive (%) | Single type (%) | Multiple types (%) |
Pb | |||
|---|---|---|---|---|---|---|---|---|---|
| ≥ 2 types | 2 types (%) | 3 types (%) | ≥ 4 types (%) | ||||||
| Swab | |||||||||
| LiPA-25 | 1,726 | 725 (42.0) | 1,001 (58.0) | 664 (38.5) | 337 (19.5) | 269 (15.6) | 55 (3.2) | 13 (0.7) | |
| Bohui-24 | 1,725 | 847 (49.1) | 878 (50.9) | 571 (33.1) | 307 (17.8) | 224 (13.0) | 70 (4.1) | 13 (0.7) | 0.587 |
| Yaneng-23 | 1,719 | 790 (46.0) | 929 (54.0) | 631 (36.7) | 298 (17.3) | 228 (13.2) | 60 (3.5) | 10 (0.6) | 0.490 |
| Tellgen-27 | 1,697 | 846 (49.9) | 851 (50.1) | 557 (32.8) | 294 (17.3) | 222 (13.1) | 56 (3.3) | 16 (0.9) | 0.706 |
| Hybribio-16 | 1,710 | 799 (46.7) | 911 (53.3) | 605 (35.4) | 306 (17.9) | 235 (13.8) | 60 (3.5) | 11 (0.6) | 1.000 |
| Hybribio-23 | 1,715 | 748 (43.6) | 967 (56.4) | 615 (35.9) | 352 (20.5) | 254 (14.8) | 77 (4.5) | 21 (1.2) | 0.221 |
| Bioperfectus-21 | 1,726 | 710 (41.1) | 1,016 (58.9) | 596 (34.6) | 420 (24.3) | 287 (16.6) | 107 (6.2) | 26 (1.5) | < 0.001 |
| Sansure-26 | 1,726 | 640 (37.1) | 1,086 (62.9) | 620 (35.9) | 466 (27.0) | 298 (17.3) | 121 (7.0) | 47 (2.7) | < 0.001 |
| Biopsy | |||||||||
| LiPA-25 | 56 | 4 (7.1) | 52 (92.9) | 52 (92.9) | 0 (0) | 0 (0) | 0 | 0 | |
| Bohui-24 | 56 | 6 (10.7) | 50 (89.3) | 50 (89.3) | 0 (0) | 0 (0) | 0 | 0 | - |
| Yaneng-23 | 56 | 4 (7.1) | 52 (92.9) | 49 (87.5) | 3 (5.4) | 3 (5.4) | 0 | 0 | 0.118 |
| Tellgen-27 | 56 | 3 (5.4) | 53 (94.6) | 51 (91.0) | 2 (3.6) | 2 (3.6) | 0 | 0 | 0.495 |
| Hybribio-16 | 56 | 3 (5.4) | 53 (94.6) | 49 (87.5) | 4 (7.1) | 4 (7.1) | 0 | 0 | 0.118 |
| Hybribio-23 | 56 | 3 (5.4) | 53 (94.6) | 47 (83.9) | 6 (10.7) | 5 (8.9) | 1 (1.8) | 0 | 0.027 |
| Bioperfectus-21 | 56 | 4 (7.1) | 52 (92.9) | 49 (87.5) | 3 (5.4) | 3 (5.4) | 0 | 0 | 0.118 |
| Sansure-26 | 56 | 2 (3.6) | 54 (96.4) | 45 (80.3) | 9 (16.1) | 9 (16.1) | 0 | 0 | 0.003 |
HPV types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66.
Chi-square test or Fisher's exact test, the difference between LiPA-25 and other 7 assays for the detection of single versus multiple genotypes (≥ 2 types).
Abbreviation: HPV, human papillomavirus.
The majority was single HPV genotype infection that ranged from 32.8% to 38.5%, while multiple genotypes infection ranged from 17.3% to 27.0% among the 8 methods, and the detection of single versus multiple genotypes was different only between Sanrue-26, Bioperfectus-21, and LiPA-25 (chi-square test: both P < 0.001) (Table 2). For 56 FFPE samples included in the study, HPV DNA was detected in 53, 50, 52, 53, 53, 53, 52, and 54 samples. All HPV DNA-positive samples were a single HPV genotype for LiPA-25 and Bohui-24. The other systems contained more or less multiple HPV genotypes (Table 2).
As stated in materials and methods, HPV genotyping results of specimens were defined as concordant, compatible, and discordant; the results are summarized in Table 3. Compared to LiPA-25, 83.8% of swab samples contained concordant genotypes and 10.3% contained compatible genotypes for Hybribio-23, 83.3% and 9.4% for Yaneng-23, 79.7% and 12.8% for Bioperfectus-21, 83.9% and 8.5% for Hybribio-16, 77.3% and 14.0% for Sansure-26, 81.2% and 8.5% for Bohui-24, and 79.5% and 8.5% for Tellgen-27, respectively. For the FFPE samples, the samples of concordant genotypes and compatible genotypes varied little (94.6%-100%) between the other 7 methods and LiPA-25.
Table 3.
Agreement between 15 HPV genotypesa detected by both LiPA-25 and other 7 assays.
| Type | Bohui-24 (%) | Yaneng-23 (%) | Tellgen-27 (%) | Hybribio-16 (%) | Hybribio-23 (%) | Bioperfectus-21 (%) | Sansure-26 (%) |
|---|---|---|---|---|---|---|---|
| Swab | n = 1,725 | n = 1,719 | n = 1,697 | n = 1,710 | n = 1,715 | n = 1,726 | n = 1,726 |
| Concordant | 1,401 (81.2) | 1,432 (83.3) | 1,349 (79.5) | 1,434 (83.9) | 1,438 (83.8) | 1,376 (79.7) | 1,334 (77.3) |
| Both negative | 700 (40.6) | 695 (40.4) | 677 (39.9) | 692 (40.5) | 682 (39.7) | 658 (38.1) | 609 (35.3) |
| Single types | 489 (28.3) | 530 (30.8) | 474 (27.9) | 522 (30.5) | 533 (31.1) | 496 (28.7) | 498 (28.9) |
| Multiple types | 212 (12.3) | 207 (12.1) | 198 (11.7) | 220 (12.9) | 223 (13.0) | 222 (12.9) | 227 (13.1) |
| Compatible | 147 (8.5) | 162 (9.4) | 144 (8.5) | 146 (8.5) | 176 (10.3) | 220 (12.8) | 241 (14.0) |
| LiPA-25 additional types | 70 (4.1) | 87 (5.1) | 64 (3.8) | 78 (4.5) | 59 (3.4) | 36 (2.1) | 28 (1.6) |
| Otherb additional types | 71 (4.1) | 71 (4.1) | 72 (4.2) | 61 (3.6) | 109 (6.4) | 179 (10.4) | 208 (12.1) |
| Both additional types | 6 (0.3) | 4 (0.2) | 8 (0.5) | 7 (0.4) | 8 (0.5) | 5 (0.3) | 5 (0.3) |
| Discordant | 177 (10.3) | 125 (7.3) | 204 (12.0) | 130 (7.6) | 101 (5.9) | 130 (7.5) | 151 (8.7) |
| LiPA-25 additional types | 147 (8.5) | 95 (5.5) | 169 (10.0) | 107 (6.2) | 66 (3.9) | 53 (3.0) | 31 (1.8) |
| Otherb additional types | 24 (1.4) | 26 (1.5) | 29 (1.7) | 20 (1.2) | 33 (1.9) | 67 (3.9) | 116 (6.7) |
| Both additional types | 6 (0.4) | 4 (0.3) | 6 (0.3) | 3 (0.2) | 2 (0.1) | 10 (0.6) | 4 (0.2) |
| Biopsy | n = 56 | n = 56 | n = 56 | n = 56 | n = 56 | n = 56 | n = 56 |
| Concordant | 53 (94.6) | 52 (92.8) | 54 (96.4) | 50 (89.3) | 48 (85.7) | 52 (92.8) | 45 (80.4) |
| Both negative | 3 (5.4) | 3 (5.4) | 3 (5.4) | 2 (3.6) | 2 (3.6) | 3 (5.4) | 2 (3.6) |
| Single types | 50 (89.2) | 49 (87.4) | 51 (91.0) | 48 (85.7) | 46 (82.1) | 49 (87.4) | 43 (76.8) |
| Multiple types | 0 (0.0) | 0 (0.0) | 0 (0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Compatible | 0 (0.0) | 3 (5.4) | 2 (3.6) | 4 (7.1) | 6 (10.7) | 3 (5.4) | 9 (16.0) |
| LiPA-25 additional types | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Otherb additional types | 0 (0.0) | 3 (5.4) | 2 (3.6) | 4 (7.1) | 6 (10.7) | 3 (5.4) | 9 (16.0) |
| Both additional types | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0) | 0 (0.0) |
| Discordant | 3 (5.4) | 1 (1.8) | 0 (0.0) | 2 (3.6) | 2 (3.6) | 1 (1.8) | 2 (3.6) |
| LiPA-25 additional types | 3 (5.4) | 1 (1.8) | 0 (0.0) | 1 (1.8) | 1 (1.8) | 1 (1.8) | 0 (0.0) |
| Otherb additional types | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.8) | 1 (1.8) | 0 (0.0) | 1 (1.8) |
| Both additional types | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.8) |
HPV types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66.
Other indicated HPV only detected by Sansure-26, Bioperfectus-21, Bohui-24, Yaneng-23, Tellgen-27, Hybribio-23, and Hybribio-16, respectively.
Abbreviation: HPV, human papillomavirus.
Subsequently, we made a comparison of individual HPV genotypes by each of the two methods. The agreement among these selected cervical swab or biopsy samples for identification by LiPA-25 and the other 7 methods is summarized in Fig. 1, Table 3, and Supplementary Tables 3-16. For most genotypes from swab specimens, including HPV types 16 and 18, the most important two genotypes, the results obtained by the two methods, including Bioperfectus-21 and LiPA-25, Yaneng-23 and LiPA-25, Hybribio-23 and LiPA-25, and Hybribio-16 and LiPA-25, were not significantly different (all, P > 0.05). To be specific, Biperfectus-21 was more sensitive than LiPA-25 for HPV types 35, 39, 56, 58, 59, and 66; Yaneng-23, Hybribio-23, and Hybribio-16 were all more sensitive than LiPA-25 for HPV types 58 and 59; whereas LiPA-25 was more sensitive for HPV types 31, 39, 52, and 66 than Yaneng-23, for HPV types 35 and 66 than Hybribio-23, and for HPV types 6, 31, 33, and 66 than Hybribio-16. When we compared Sansure-26 and LiPA-25, for most genotypes, including HPV types 16 and 18, Sansure-26 was more sensitive than LiPA25. The results for the HPV types 31, 35, 45, and 59 were not significantly different. Regarding Bohui-24 and LiPA-25, LiPA-25 was more sensitive for most genotypes, while Bohui-24 was more sensitive for only HPV type 59. The HPV types 16, 18, 33, 45, 56, and 58 were no statistical evidence of an imbalance. As for Tellgen-27 and LiPA-25, LiPA-25 was more sensitive for HPV types 11, 31, 33, 45, 51, 52, and 66, while Tellgen-27 was more sensitive for HPV types 58 and 59. The results for the remaining HPV types were not significantly different. In 56 biopsy samples, we did not detect all of the 15 individual HPV types, and HPV 11, 33, 39, and 66 were detected by the 8 systems. Specifically, over 40 samples were detected HPY type 16 or 18 by the 8 systems, respectively, and the other HPV types were all present in less than 4 samples.
Almost perfect agreement for the 15 HPV-combined detection in swab specimens was found in LiPA-25 and the other 7 systems (Table 4), i.e., Sansure-26 (Cohen's κ = 0.826), Bioperfectus-21 (κ = 0.848), Bohui-24 (κ = 0.847), Yaneng-23 (κ = 0.871), Tellgen-27 (κ = 0.831), Hybribio-23 (κ = 0.879), and Hybribio-16 (κ = 0.877). To be specific, the lowest kappa value was HPV 52 for Sansure-26, and Hybribio-16, and HPV31 for other 5 systems (Supplementary Tables 3-9). In biopsy specimens for 15 HPV combined detection, similar results were obtained (all, κ > 0.897).
Table 4.
Overall agreement between SPF10 LiPA-25 and other 7 assays in detecting 15 HPV genotypesa in cervical swab and biopsy specimens.
| Methods | No. of samples with LiPA-25 and other assayb result of: |
Ppos | Pneg | Kappa value (95% CI) | Pc | |||
|---|---|---|---|---|---|---|---|---|
| +/+ | +/- | -/+ | -/- | |||||
| Swab | ||||||||
| Bohui-24 | 1,156 | 266 | 127 | 24,326 | 0.855 | 0.992 | 0.847 (0.839-0.855) | < 0.001 |
| Yaneng-23 | 1,197 | 222 | 112 | 24,254 | 0.878 | 0.993 | 0.871 (0.864-0.878) | < 0.001 |
| Tellgen-27 | 1,110 | 298 | 126 | 23,921 | 0.840 | 0.991 | 0.831 (0.823-0.839) | < 0.001 |
| Hybribio-16 | 1,201 | 216 | 100 | 24,133 | 0.884 | 0.993 | 0.877 (0.870-0.884) | < 0.001 |
| Hybribio-23 | 1,268 | 150 | 178 | 24,129 | 0.885 | 0.993 | 0.879 (0.872-0.886) | 0.136 |
| Bioperfectus-21 | 1,294 | 128 | 306 | 24,162 | 0.856 | 0.991 | 0.848 (0.841-0.855) | < 0.001 |
| Sansure-26 | 1,344 | 78 | 447 | 24,021 | 0.837 | 0.989 | 0.826 (0.819-0.833) | < 0.001 |
| Biopsy | ||||||||
| Bohui-24 | 50 | 2 | 0 | 788 | 0.980 | 0.999 | 0.979 (0.964-0.994) | 0.500 |
| Yaneng-23 | 52 | 0 | 3 | 785 | 0.972 | 0.998 | 0.970 (0.953-0.987) | 0.250 |
| Tellgen-27 | 52 | 0 | 3 | 785 | 0.972 | 0.998 | 0.970 (0.953-0.987) | 0.250 |
| Hybribio-16 | 52 | 0 | 5 | 783 | 0.954 | 0.997 | 0.951 (0.929-0.973) | 0.063 |
| Hybribio-23 | 51 | 1 | 9 | 779 | 0.911 | 0.994 | 0.904 (0.874-0.934) | 0.021 |
| Bioperfectus-21 | 52 | 0 | 3 | 785 | 0.972 | 0.998 | 0.970 (0.953-0.987) | 0.250 |
| Sansure-26 | 52 | 0 | 11 | 777 | 0.904 | 0.993 | 0.897 (0.867-0.928) | 0.001 |
HPV types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 66.
Other assay indicated LiPA-25, Sansure-26, Bioperfectus-21, Bohui-24, Yaneng-23, Tellgen-27, Hybribio-23 and Hybribio-16, respectively.
McNemar test.
Abbreviation: CI, confidence interval; HPV, human papillomavirus; Ppos, proportion of positive agreement; Pneg, proportion of negative agreement.
The highest Ppos for the 15 types combined in swab specimens was Hybribio-23 with 0.885 (ranged from 0.805 for HPV31 to 0.939 for HPV18), and followed by Hybribio-16 with 0.884 (0.790 for HPV52 to 0.927 for HPV18), Yaneng-23 with 0.878 (0.729 for HPV39 to 0.989 for HPV35), Bioperfectus-21 with 0.856 (0.766 for HPV31 to 0.950 for HPV11), Bohui-24 with 0.855 (0.795 for HPV31 to 0.935 for HPV11), Tellgen-27 with 0.840 (0.739 for HPV31 to 0.894 for HPV45), and Sansure-26 with 0.837 (0.754 for HPV31 to 0.933 for HPV18). In contrast, the variation of Pneg between the two assays (other 7 methods and LiPA-25) was very small (ranged from 0.968 to 0.999), and the least of Pneg was all HPV 52 for the 7 systems. In biopsy specimens, the range of Ppos between the two assays (other 7 methods and LiPA-25) was 0.904 for Sansure-26 to 0.980 for Bohui-24, and the range of Pneg was 0.993 for Sansure-26 to 0.999 for Bohui-24 as well.
As mentioned above, overall agreement rates for the detection of 15 HPV types were high (all, κ > 0.826); however, it is clear that this agreement resulted from the agreement of HPV-negative specimens, as shown through the high Pneg rates. The observed lower proportions of positive agreement index that there are discrepancies in the different assays’ abilities to determine type-specific HPV positives, which suggests differences in assay sensitivity.
In total, LiPA-25 detected 1,422 HPV genotypes (15 types-combined) in swab specimens, which was significantly lower than 1,791 and 1,600 genotypes found by sansure-26 (P < 0.001) and Bioperfectus-21 (P < 0.001) and was significantly higher than 1,283 by Bohui-24 (P < 0.001), 1,309 by Yaneng-23 (P < 0.001), 1,236 by Tellgen-27 (P < 0.001), and 1,301 by Hybribio-16 (P < 0.001), respectively (Table 4). Although the total of HPV genotypes (1,446) detected by Hybribio-23 was higher than those of LiPA-25, there was no significant difference (P = 0.136, Table 4). Regarding biopsy, multiple HPV infection was not detected by LiPA-25, and the number of specimens was relatively few. A significant difference was found between Sansure-26 and LiPA-25, and between Hybribio-23 and LiPA-25.
3.2. Detection by TS16 and TS18 PCR/DEIA
For improving the detection rates, we conducted the detection of TS16 and TS18 PCR/DEIA for the positive SPF10 DEIA samples. For swab samples, the number of HPV16 positive sample detected by SPF10 LiPA-25 was 100, and the union of two sets (HPV16 positive of LiPA-25 and TS16 DEIA positive) was 117 (Supplementary Tables 3-9). The number of HPV18-positive sample was 84, which is only 2 less than the union of two sets (HPV18 positive for LiPA-25 and TS18 DEIA positive) (Supplementary Tables 3-9). Regarding biopsy, the result of LiPA-25 was identical with that of TS16 or TS18 DEIA.
4. Discussion
Since the HPV persistent infection is a surrogate virological endpoint in vaccine trials, it is vital to use reliable and precise testing methods for HPV genotypes to evaluate the efficacy of candidate HPV vaccines.27 The accuracy and precision of HPV genotypes detection on cervical specimens (cervical swab and biopsy specimens) depend on the type of the molecular technology platform used. In this study, 4 systems of hybridization (2 membrane-based, 1 microfluidics + membrane-based, and 1 bead-based) and 3 systems of real-time fluorescence PCR were compared with the LiPA-25 system for identification of the commonly targeted 15 HPV genotypes. The results indicated a high concordance, although the other 7 systems were executed under suboptimal conditions. Briefly, in 1,726 swab specimens, compared with LiPA-25, 88.0% for Sansure-26 to 94.1% for Hybribio-23 of swab samples contained concordant and compatible HPV genotypes. Similar results were obtained for 56 biopsy specimens, for which >94.6% of samples for all 7 systems contained concordant and compatible genotypes. Moreover, these systems showed excellent strength of overall agreements for 15 genotypes detection in swab (all, κ > 0.829) or biopsy specimens (all, κ > 0.897).
For broad-spectrum and multiplex type-specific PCR assays, a competition can occur between multiple HPV genotypes within a test, especially, when some HPV genotypes are present at relatively low concentrations, while other types are present at high levels. Due to this competition effect, these approaches may underestimate the presence of HPV genotypes. It is also likely that the competition from the multiplex type-specific PCR approach is weaker than that from broad-spectrum PCR assays. In both swab and biopsy specimens, the total amount of multiplex type-specific PCR assays (e.g., Sansure-26, Bioperfectus-21, and Hybribio-23) for 15 HPV genotypes was more than that of the broad-spectrum PCR assay (e.g., LiPA-25), which suggested the former was more sensitive. Moreover, the three systems could detect more multiple genotypes in individual samples than LiPA-25. Another explanation is that based on the real-time PCR method, the sample is divided into multi-reaction tubes, which could reduce competitive inhibition of detection of multiple HPV infections. These results demonstrated using a broad-spectrum PCR-based approach can be circumvented by using a multiplex type-specific PCR assay for PCR competition among multiple HPV genotypes.
In contrast, although LiPA-25 is a broad-spectrum PCR assay as Hybribio-16, Yaneng-23, Bohui-24, and Tellgen-27, LiPA-25 was more sensitive than the others. One probable reason is that the SPF10 primer set amplifying a 65-bp region is shorter than that of other primer sets, such as GP5+/6+ and MY09/11, due to the fact that a shorter amplification product was thought to be more analytically sensitive for HPV detection. There might also be some false-positive results due to lack of an internal control in LiPA-25 system, while all other systems have an internal control with human DNA (housekeeping gene).
It is known that each PCR primer set has a marginal preference for different HPV types, which is more likely to be amplified. LiPA-25 can detect more HPV 31 and 52 and less HPV 58 or 59 (Fig 1 and Supplementary Table 3-9) than most of the other 7 methods, although no statistical evidence of an imbalance between Bioperfectus-21, Hybribio-23, and LiPA-25 was found for HPV 31 and 52. Previous studies reported similar results.27,36 For example, van Alewijk et al.36 tested 400 cervical scraping specimens using LiPA-25 and PGMY Line Blot Assay (LBA). LiPA-25 significantly appeared more sensitive for the detection of HPV 31 (P < 0.05) and 52 (P < 0.05), less sensitive for HPV 59 (P < 0.05) than PGMY LBA. In another study on the comparison between LiPA-25 and MPTS123 system, a E6-based multiplex type-specific system, for 860 swab samples, MPTS123 system was significantly more sensitive for HPV 58 (P < 0.05) and HPV 59 (P < 0.05).27 Multiple HPV vaccines, which can prevent cervical cancer that results from different HPV types, have been launched and approved for use in individuals with different age in the world. Studies have shown that HPV subtypes have significant age and regional specificities.37, 38, 39, 40 In 2013, the working group at the IARC/United States National Cancer Institute (NCI) Expert Meeting recommended using a virological end-point (persistent HPV infection for 6 months or longer), rather than a disease end-point such as CIN2+, as the primary end-point for some future clinical efficacy trials of vaccine.41 In United States or Europe, the prevalent HPV types were 16, 18, 45, 31, and 33. The 5 common high-risk types with infection rates were 16, 52, 58, 53, and 18 in Chinese mainland.39 In addition, in China, the most five common HPV types in CIN1 were HPV 52, 16, 58, 18, and 53,42 in CN2/3 were HPV 16, 58, 52, 33, and 31.42 Considering that the high prevalence of HPV 52 and 58 is just below HPV 16, both in the general population and in patients with CIN2+, the Hybribio-23 and bioperfectus-21 with high-sensitive 52 and 58 are suitable for the novel vaccine, especially applicable to the Chinese population.
As described in a previous study,43 FFPE biopsy tissue specimens pose a critical challenge for DNA extraction and subsequent HPV identification. The efficiency of PCR amplification is inversely correlated to the length of the amplimer produced. The amplimer lengths of the other 7 systems area from 120 bp to 450 bp, longer than that of SPF10 (65 bp). However, the other 7 systems detected more types, except Bohui-24. Moreover, the use of type-specific primer pairs increased the detection of HPV DNA from FFPE biopsy specimens.43 The Hybriobio-23, Sansure-26, and Bioperfectus-21, three systems with multiplex type-specific primer-added multi-detection tubes, were more sensitive for the 15 HPV types than LiPA-25.
Either system could be used as a stand-alone system to detect and identify HPV genotypes. To improve the detection rate of types, the combination of SPF10 LiPA-25 and the type-specific PCR DEIA systems for HPV16 and HPV18 is a well-established methodology for HPV16 & 18 vaccine studies.14,15,19 Consequently, it also increased the workload and the cost associated with HPV genotyping. In our study, we compared the combination of LiPA-25 plus TS16 or TS18 PCR/DEIA with other 7 systems, respectively, and the combined agreement rate of HPV16 moderately increased for all 7 systems. In contrast, the combined agreement rate of HPV 18 for Bohui-24 or Tellgen-27 decreased slightly, while for the other systems, there was a marginal rise, comparing the results of HPV 18 for only LiPA-25.
There are also disadvantages and advantages of these HPV genotyping systems. In general, the sample size is challenging for HPV genotyping because more participants need to be recruited to evaluate the effects for more values vaccine. Therefore, it is necessary to use a high-throughput and automatic or semi-automatic method for HPV genotyping. Although the real-time PCR methods are more sensitive for HPV genotyping, the workload is 6-8 folds as much as hybridization methods because the samples need to be parallelly divided into 6-8 detection tubes. The microarray hybridization from Hybribio-16 utilized a flow-through hybridization technique by actively directing the targeting molecules toward the immobilized probes within the membrane fibers. Compared with conventional reverse dot hybridization, the method makes the DNA into the pore of hybrid membrane by diversion, which greatly improves the speed and efficiency of hybridization. The nucleic acid chip detector (BHF-VI) from Bohui-24 was fully automatic, and the swab samples were only added to the wells from HPV detection chip. The three processes, including target DNA extraction, PCR amplification, and reverse dot hybridization, are carried out under the drive of a micropump. After about 4 h, the software in instrument can automatically report the results of detected sample. Therefore, besides the accuracy of HPV genotypes detection, workload, reaction time, and other factors should be taken into consideration.
There are several strengths and limitations in the study. The most important strength was that this was a head-to-head and large-scale study covering detection of all the high-risk HPV and the most important two low-risk HPV 6 and HPV 11 in two specimens, including exfoliated cervical cells and FFPE samples. The primer targets regions from the 8 selected systems not only include the later region but also the early region. In addition, all these systems except LiPA-25 included the housekeeping gene as an internal control, which could reduce sampling errors and false negative detection. There are also limitations. First, the panel size of FFPE biopsy specimens was much smaller than that of swabs, and not every HPV genotype was included. Moreover, the composition of the panel of 1,726 swab samples was based on the results of SPF10 LiPA-25 system obtained 3 to 5 years ago. Although all the samples were stored at -80°C, it might have a negative influence on the detection of HPV genotypes due to DNA degeneration.
In conclusion, analytically sensitive and type-specific measures of HPV infections are vital for continually monitoring of HPV infection and determination of the causality of precancer. Detection of the 15 HPV types common to LiPA-25 without a human control primer and other 7 systems with housekeeping gene primers was similar in swab and biopsy specimens, although there were more or less different in the 15 HPV types in comparison. Considering the prevalence of different types, e.g., HPV 31, 52, and 58, in population, Hybribio-23 is highly suitable for studies on multiple-value vaccine efficacy, genotype surveillance, and disease association. We also need to consider the operating procedure and workload of the HPV genotyping method for vaccine and epidemiological studies. This study contributes to our understanding of the performance characteristics, practicability, and comparability of the 8 systems of HPV genotyping.
Acknowledgments
Declaration of competing interest
The authors declare that they have no conflict of interests.
Ethics statement
This study was performed in accordance with the recommendations of the Declaration of Helsinki and approved by the Institutional Review Board of Cancer Hospital, Chinese Academy of Medical Sciences (No. 20/243-2439).
Acknowledgments
This work was supported by the CAMS Innovation Fund for Medical Sciences (grant number 2021-I2M-1-004) and the National Natural Science Foundation of China (grant number 81973136).
Author contributions
W.C., J.Y., Y.Q., C.Z., and S.X. designed the study. J.Y., S.C., D.L., Y.T., F.H., Z.Z., T.Z., Z.S., Y.L., S.W., Y.L., S.P., and L.L. managed the data. J.Y. conducted the statistical analysis and drafted the paper. W.C., Y.T., S.X., C.Z., and Y.Q. reviewed the paper. All of the authors commented on the draft and decided to submission.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jncc.2022.06.003.
Contributor Information
Chuntao Zhang, Email: zhangct2@126.com.
Wen Chen, Email: chenwen@cicams.ac.cn.
Appendix. Supplementary materials
References
- 1.Zhang SW, Sun KX, Zheng RS, et al. Cancer incidence and mortality in China, 2015. Journal of National Cancer Center. 2021;1(1):2–11. doi: 10.1016/j.jncc.2020.12.001. [DOI] [Google Scholar]
- 2.Zhao F, Qiao Y. Cervical cancer prevention in China: a key to cancer control. Lancet. 2019;393(10175):969–970. doi: 10.1016/S0140-6736(18)32849-6. [DOI] [PubMed] [Google Scholar]
- 3.Choi YJ, Park JS. Clinical significance of human papillomavirus genotyping. J Gynecol Oncol. 2016;27(2):e21. doi: 10.3802/jgo.2016.27.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sundstrom K, Eloranta S, Sparen P, et al. Prospective study of human papillomavirus (HPV) types, HPV persistence, and risk of squamous cell carcinoma of the cervix. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2469–2478. doi: 10.1158/1055-9965.EPI-10-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kjaer SK, van den Brule AJ, Paull G, et al. Type specific persistence of high risk human papillomavirus (HPV) as indicator of high grade cervical squamous intraepithelial lesions in young women: population based prospective follow up study. BMJ. 2002;325(7364):572. doi: 10.1136/bmj.325.7364.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11(11):1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 7.Stanley M. Pathology and epidemiology of HPV infection in females. Gynecol Oncol. 2010;117(2 Suppl):S5–10. doi: 10.1016/j.ygyno.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 8.Small W, Jr., Bacon MA, Bajaj A, et al. Cervical cancer: A global health crisis. Cancer. 2017;123(13):2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 9.Latsuzbaia A, Arbyn M, Tapp J, et al. Effectiveness of bivalent and quadrivalent human papillomavirus vaccination in Luxembourg. Cancer Epidemiol. 2019;63 doi: 10.1016/j.canep.2019.101593. [DOI] [PubMed] [Google Scholar]
- 10.Jacot-Guillarmod M, Pasquier J, Greub G, et al. Impact of HPV vaccination with Gardasil(R) in Switzerland. BMC Infect Dis. 2017;17(1):790. doi: 10.1186/s12879-017-2867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latsuzbaia A, Tapp J, Nguyen T, et al. Analytical performance evaluation of Anyplex II HPV28 and Euroarray HPV for genotyping of cervical samples. Diagn Microbiol Infect Dis. 2016;85(3):318–322. doi: 10.1016/j.diagmicrobio.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 12.van Hamont D, van Ham MA, Bakkers JM, et al. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the roche linear array HPV genotyping test. J Clin Microbiol. 2006;44(9):3122–3129. doi: 10.1128/JCM.00517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castle PE, Porras C, Quint WG, et al. Comparison of two PCR-based human papillomavirus genotyping methods. J Clin Microbiol. 2008;46(10):3437–3445. doi: 10.1128/JCM.00620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrero R, Wacholder S, Rodriguez AC, et al. Prevention of persistent human papillomavirus infection by an HPV16/18 vaccine: a community-based randomized clinical trial in Guanacaste, Costa Rica. Cancer Discov. 2011;1(5):408–419. doi: 10.1158/2159-8290.CD-11-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreimer AR, Gonzalez P, Katki HA, et al. Efficacy of a bivalent HPV 16/18 vaccine against anal HPV 16/18 infection among young women: a nested analysis within the Costa Rica Vaccine Trial. Lancet Oncol. 2011;12(9):862–870. doi: 10.1016/S1470-2045(11)70213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehtinen M, Paavonen J, Wheeler CM, et al. Overall efficacy of HPV-16/18 AS04-adjuvanted vaccine against grade 3 or greater cervical intraepithelial neoplasia: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012;13(1):89–99. doi: 10.1016/S1470-2045(11)70286-8. [DOI] [PubMed] [Google Scholar]
- 17.Wheeler CM, Castellsague X, Garland SM, et al. Cross-protective efficacy of HPV-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by non-vaccine oncogenic HPV types: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012;13(1):100–110. doi: 10.1016/S1470-2045(11)70287-X. [DOI] [PubMed] [Google Scholar]
- 18.Hildesheim A, Wacholder S, Catteau G, et al. Efficacy of the HPV-16/18 vaccine: final according to protocol results from the blinded phase of the randomized Costa Rica HPV-16/18 vaccine trial. Vaccine. 2014;32(39):5087–5097. doi: 10.1016/j.vaccine.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao YL, Wu T, Li RC, et al. Efficacy, Safety, and Immunogenicity of an Escherichia coli-Produced Bivalent Human Papillomavirus Vaccine: An Interim Analysis of a Randomized Clinical Trial. J Natl Cancer Inst. 2020;112(2):145–153. doi: 10.1093/jnci/djz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu FC, Chen W, Hu YM, et al. Efficacy, immunogenicity and safety of the HPV-16/18 AS04-adjuvanted vaccine in healthy Chinese women aged 18-25 years: results from a randomized controlled trial. Int J Cancer. 2014;135(11):2612–2622. doi: 10.1002/ijc.28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alemany L, Saunier M, Tinoco L, et al. Large contribution of human papillomavirus in vaginal neoplastic lesions: a worldwide study in 597 samples. Eur J Cancer. 2014;50(16):2846–2854. doi: 10.1016/j.ejca.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 22.Schmeink CE, Massuger LF, Lenselink CH, et al. Prospective follow-up of 2,065 young unscreened women to study human papillomavirus incidence and clearance. Int J Cancer. 2013;133(1):172–181. doi: 10.1002/ijc.27986. [DOI] [PubMed] [Google Scholar]
- 23.Geraets DT, Struijk L, Kleter B, et al. The original SPF10 LiPA25 algorithm is more sensitive and suitable for epidemiologic HPV research than the SPF10 INNO-LiPA Extra. J Virol Methods. 2015;215-216:22–29. doi: 10.1016/j.jviromet.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Sun Z, Zhang R, Liu Z, et al. Development of a fluorescence-based multiplex genotyping method for simultaneous determination of human papillomavirus infections and viral loads. BMC Cancer. 2015;15:860. doi: 10.1186/s12885-015-1874-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Else EA, Swoyer R, Zhang Y, et al. Comparison of real-time multiplex human papillomavirus (HPV) PCR assays with INNO-LiPA HPV genotyping extra assay. J Clin Microbiol. 2011;49(5):1907–1912. doi: 10.1128/JCM.00236-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iftner T, Germ L, Swoyer R, et al. Study comparing human papillomavirus (HPV) real-time multiplex PCR and Hybrid Capture II INNO-LiPA v2 HPV genotyping PCR assays. J Clin Microbiol. 2009;47(7):2106–2113. doi: 10.1128/JCM.01907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Alewijk D, Kleter B, Vent M, et al. A human papilloma virus testing algorithm comprising a combination of the L1 broad-spectrum SPF10 PCR assay and a novel E6 high-risk multiplex type-specific genotyping PCR assay. J Clin Microbiol. 2013;51(4):1171–1178. doi: 10.1128/JCM.02831-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estrade C, Sahli R. Comparison of Seegene Anyplex II HPV28 with the PGMY-CHUV assay for human papillomavirus genotyping. J Clin Microbiol. 2014;52(2):607–612. doi: 10.1128/JCM.02749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hesselink AT, Sahli R, Berkhof J, et al. Clinical validation of Anyplex II HPV HR Detection according to the guidelines for HPV test requirements for cervical cancer screening. J Clin Virol. 2016;76:36–39. doi: 10.1016/j.jcv.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Sun P, Song Y, Ruan G, et al. Clinical validation of the PCR-reverse dot blot human papillomavirus genotyping test in cervical lesions from Chinese women in the Fujian province: a hospital-based population study. J Gynecol Oncol. 2017;28(5):e50. doi: 10.3802/jgo.2017.28.e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang HL, Zhu HH, Zhou LF, et al. Genotyping of human papillomavirus in cervical lesions by L1 consensus PCR and the Luminex xMAP system. J Med Microbiol. 2006;55(Pt 6):715–720. doi: 10.1099/jmm.0.46493-0. [DOI] [PubMed] [Google Scholar]
- 32.Zhao PY, Jiang HC, Li Y, et al. Comparison of the cervista HPV HR test and luminex XMAP technology for the diagnosis of cervical intraepithelial neoplasia. Eur J Obstet Gynecol Reprod Biol. 2017;214:150–155. doi: 10.1016/j.ejogrb.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Wagner S, Roberson D, Boland J, et al. Evaluation of TypeSeq, a Novel High-Throughput, Low-Cost, Next-Generation Sequencing-Based Assay for Detection of 51 Human Papillomavirus Genotypes. J Infect Dis. 2019;220(10):1609–1619. doi: 10.1093/infdis/jiz324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y, Meng Y, Yang S, et al. Screening of Cervical Cancer with Self-Collected Cervical Samples and Next-Generation Sequencing. Dis Markers. 2018;2018 doi: 10.1155/2018/4826547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjalma WA, Fiander A, Reich O, et al. Differences in human papillomavirus type distribution in high-grade cervical intraepithelial neoplasia and invasive cervical cancer in Europe. Int J Cancer. 2013;132(4):854–867. doi: 10.1002/ijc.27713. [DOI] [PubMed] [Google Scholar]
- 36.van Doorn LJ, Quint W, Kleter B, et al. Genotyping of human papillomavirus in liquid cytology cervical specimens by the PGMY line blot assay and the SPF(10) line probe assay. J Clin Microbiol. 2002;40(3):979–983. doi: 10.1128/jcm.40.3.979-983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou HL, Zhang W, Zhang CJ, et al. Prevalence and distribution of human papillomavirus genotypes in Chinese women between 1991 and 2016: A systematic review. J Infect. 2018;76(6):522–528. doi: 10.1016/j.jinf.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Chan PK, Ho WC, Chan MC, et al. Meta-analysis on prevalence and attribution of human papillomavirus types 52 and 58 in cervical neoplasia worldwide. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li K, Li Q, Song L, et al. The distribution and prevalence of human papillomavirus in women in mainland China. Cancer. 2019;125(7):1030–1037. doi: 10.1002/cncr.32003. [DOI] [PubMed] [Google Scholar]
- 40.Clifford GM, Smith JS, Plummer M, et al. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88(1):63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyon (FR); 2014. Primary End-points for Prophylactic HPV Vaccine Trials. [PubMed] [Google Scholar]
- 42.Zhang J, Cheng K, Wang Z. Prevalence and distribution of human papillomavirus genotypes in cervical intraepithelial neoplasia in China: a meta-analysis. Arch Gynecol Obstet. 2020;302(6):1329–1337. doi: 10.1007/s00404-020-05787-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baay MF, Quint WG, Koudstaal J, et al. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol. 1996;34(3):745–747. doi: 10.1128/jcm.34.3.745-747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.