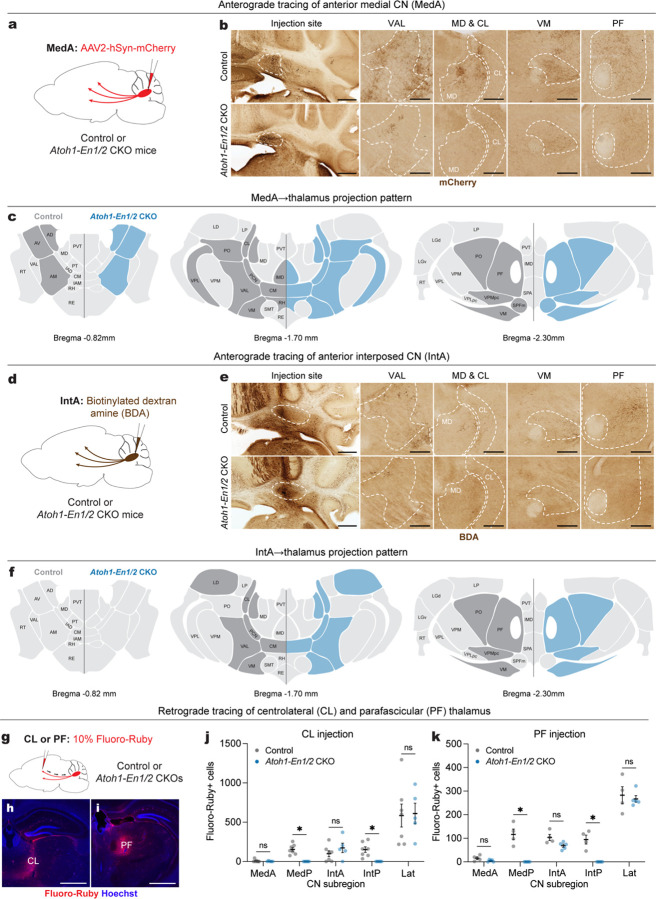
Fig. 7 |. Atoh1-En1/2 CKOs have reduced cerebellothalamic projections, but no ectopic cerebellothalamic projections.
a, Schematic of anterograde tracing of MedA CN cells in adult Atoh1-En1/2 CKOs and littermate controls.
b, Representative images of coronal sections showing injection site and mCherry+ axon terminals (brown) in various thalamic regions from an Atoh1-En1/2 CKO and littermate control. Scale bar: injection site = 1 mm and mCherry images = 250 um.
c, Summary of mCherry+ axon terminals observed in thalamic nuclei of Atoh1-En1/2 CKOs (blue) versus littermate controls (dark grey) on three representative coronal planes adapted from Allen Brain Atlas. Blue indicates reduced density.
d, Schematic of anterograde tracing of anterior interposed CN (IntA) cells in adult Atoh1-En1/2 CKOs and littermate controls.
e, Representative images of coronal sections showing injection site and biotinylated dextran amine (BDA)+ axon terminals (brown) in various thalamic regions from an Atoh1-En1/2 CKO and littermate control. Scale bar: injection site = 1 mm and BDA images = 250 um.
f, Summary of BDA+ axon terminals observed in thalamic nuclei of Atoh1-En1/2 CKOs (blue) and littermate controls (dark grey) versus on three representative coronal planes adapted from Allen Brain Atlas. Blue indicates reduced density.
g, (top) Schematic of retrograde tracing in adult Atoh1-En1/2 CKOs and littermate controls.
h,i, Representative images of the injection site of Fluoro-Ruby (red) and Hoechst (blue) in centrolateral thalamus (CL, h) and parafascicular thalamus (PF, i). Scale bars = 1 mm.
j, Quantification of Fluoro-Ruby+ cells in CN subregions that are retrogradely labeled from CL injection. Multiple Mann-Whitney U tests with Holm-Šídák correction for effect of genotype on MedP (U = 0, P = 0.0126) and IntP (U = 0, P = 0.0126), but not on MedA (U = 15.5, P = 0.9400), IntA (U = 11, P = 0.6882), and Lat (U = 15, P = 0.9400).
k, Quantification of Fluoro-Ruby+ cells in CN subregions that are retrogradely labeled from PF injection. Multiple Mann-Whitney U tests with Holm-Šídák correction for effect of genotype on MedP (U = 0, P = 0.0390) and IntP (U = 0, P = 0.0390), but not on MedA (U = 3.5, P = 0.2516), IntA (U = 1, P = 0.0922), and Lat (U = 10, P > 0.9999).
Abbreviations: AD=Anterodorsal nucleus; AM=Anteromedial nucleus; AV=Anteroventral nucleus of thalamus; CL=Central lateral nucleus; CM=Central medial nucleus; IAD=Interanterodorsal nucleus; IAM=Interanteromedial nucleus; IMD=Intermediodorsal nucleus; LD=Lateral dorsal nucleus of thalamus; LGv=Ventral part of the lateral geniculate complex; LP=Lateral posterior nucleus; MD=Mediodorsal nucleus of thalamus; PCN=Paracentral nucleus; PF=Parafascicular nucleus; PO=Posterior complex; PT=Parataenial nucleus; PVT=Paraventricular nucleus; RE=Nucleus of reuniens; RH=Rhomboid nucleus; RT=Reticular nucleus; SMT=Submedial nucleus; SPFm=Subparafascicular nucleus, magnocellular part; VAL=Ventral anterior-lateral complex; VM=Ventral medial nucleus; LGd=Dorsal part of the lateral geniculate complex; VPM=Ventral posteromedial nucleus; VPL=Ventral posterolateral nucleus; SPA=Subparafascicular area; VPMpc=Ventral posteromedial nucleus, parvicellular part; VPLpc=Ventral posterolateral nucleus, parvicellular part. ns, not significant: P ≥ 0.05. Data are presented as mean values ± SEM.