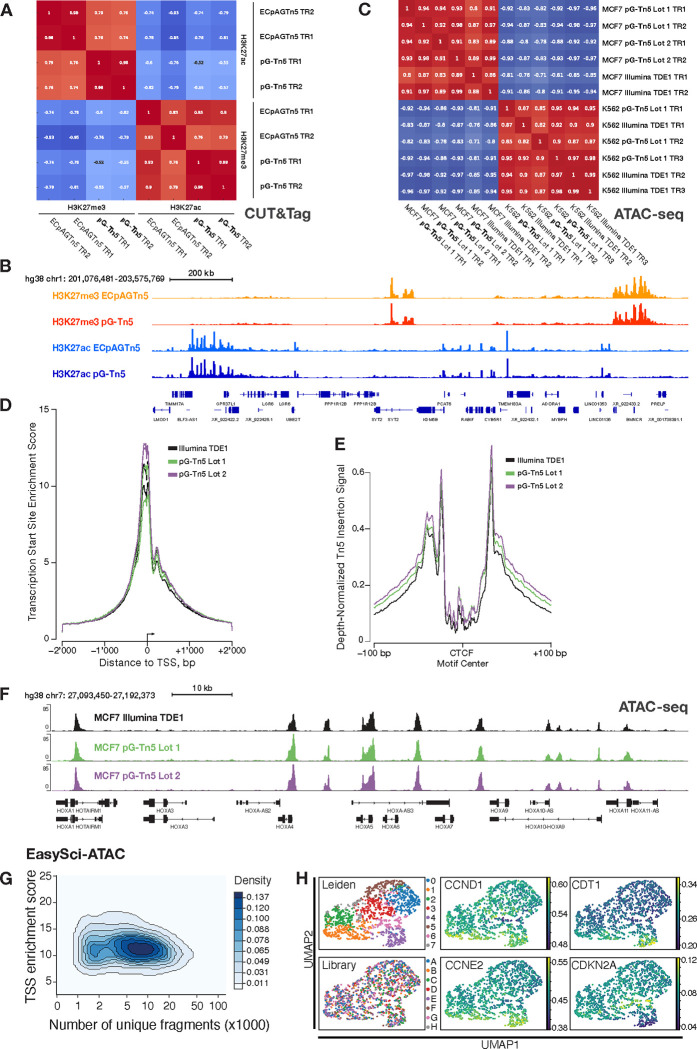
Figure 2: Application and validation of pG-Tn5 in CUT&Tag, ATAC-seq and EasySci-ATAC assays.
A. Spearman rank correlation of anti-H3K27me3 and anti-H3K27ac CUT&Tag signal under common peaks generated using pG-Tn5 and Epicypher pAG-Tn5 (ECpAGTn5) enzyme (Cat #15–1017) in human MCF7 cells. B. Genome track visualization of combined two technical replicates of above conditions, as in panel A. H3K27me3 and H3K27ac signal from pG-Tn5 and pAG-Tn5 experiments plotted on same sequencing-depth scaled y-axes, respectively. C. Spearman rank correlation of signal under common peaks, generated using ATAC-seq protocol generated libraries from human K562 and MCF7 cells using pG-Tn5 and Illumina TDE1, using matched transposome volumes. D. Transcription start site enrichment score of ATAC-seq signal in human MCF7 cells, comparison of two purification lots of pG-Tn5 and Illumina TDE1 enzyme, two technical replicates each. E. Sequencing-depth normalized Tn5 insertion signal around occupied CTCF sites in human MCF7 cells for ATAC-seq signal from pG-Tn5 and Illumina TDE1, as above in panel D. F. Genome track visualization of ATAC-seq signal from pG-Tn5 and Illumina TDE1 generated libraries, combined two technical replicates of above conditions, as in panel D and E. G. Single-cell accessible chromatin mapping using pG-Tn5 loaded with six unique barcoded adapter combinations, using the EasySci-ATAC protocol in human MCF7 cells. TSS enrichment scores for 2’672 single cells, majority of the cells showing score >10. H. Leiden clustering annotations (0–7) of EasySci-ATAC data from cycling human MCF7 cells from eight library replicates (A-H), showing no library-based bias in clustering. Signal values from several cell cycle-associated genes is shown.