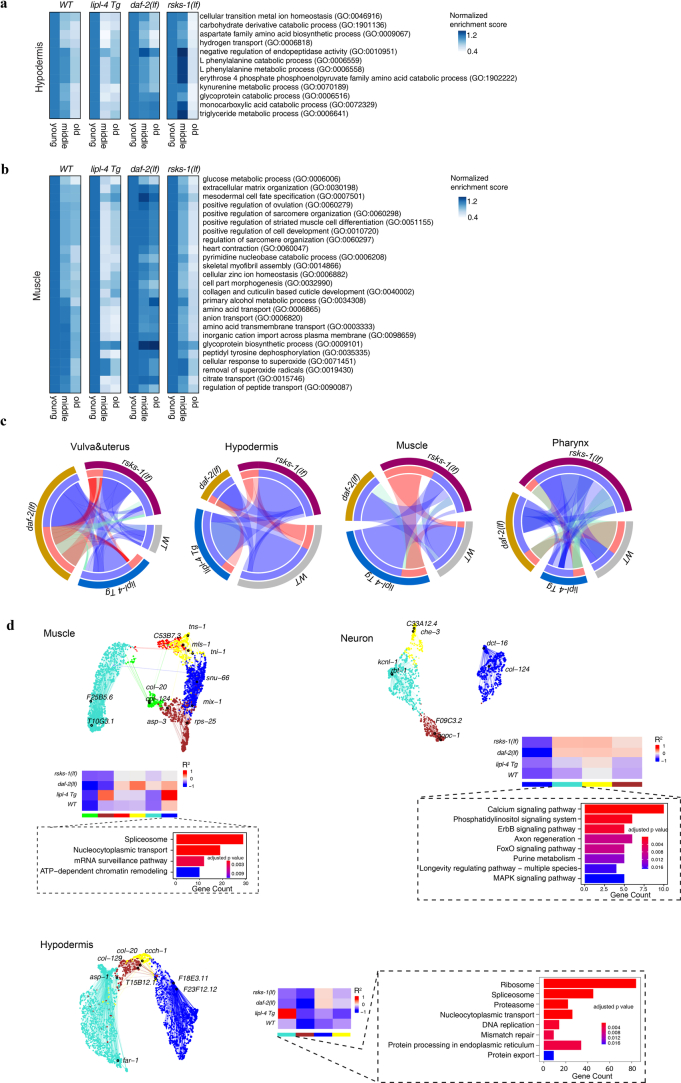
Extended Data Fig. 6. Age-related molecular regulations of tissue-specific aging by different pro-longevity mechanisms.
a and b, Heatmaps show that tissue-specific significant age-related Gene Ontology (GO) terms for hypodermis (a) and muscle (b) exhibit distinct patterns of changes in the three long-lived strains. The enrichment scores for each term were normalized to the young group. c, Circos plots showing conserved co-expression modules (Fisher’s exact test, P < 0.01) that are significantly correlated with aging (Pearson’s correlation, P < 0.001 and R2 > 0.2) in vulva and uterus, hypodermis, muscle, and pharynx between different genotypes. Blue ribbons connected conserved models that were both negatively correlated with aging in different genotypes, red ribbons connected conserved models that were both positively correlated with aging, and green ribbons connected conserved models that were oppositely correlated with aging in different genotypes. d, UMAP visualization of the consensus co-expression network for aging-related modules (Pearson’s correlation, P < 0.001 and R2 > 0.2) in muscle, neurons and hypodermis. Dots represented genes and are colored by the module they belonged to. Edges represent co-expression between genes (upper). Lower panel are heatmaps representing correlations between consensus co-expression modules and aging. Gene modules demonstrating significant correlation with aging (Pearson’s correlation, P < 0.001 and R2 > 0.2) in at least one genotype are shown. Bar graphs enclosed in dashed boxes represent KEGG pathways that are significantly enriched (Fisher’s exact test, Benjamini–Hochberg adjusted P < 0.01) for the blue module in the muscle, turquoise module in neurons, and turquoise module in the hypodermis.