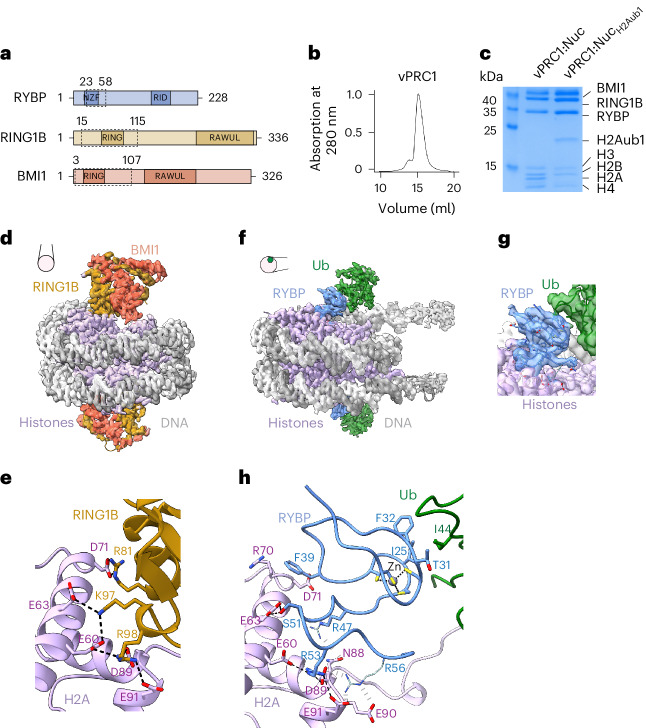
Fig. 1. vPRC1 binds unmodified nucleosomes via RING1B:BMI1 and H2Aub1-modified nucleosomes via RYBP.
a, Domain organization of the vPRC1 subunits with the NZF and Ring1b-interacting domain (RID) in RYBP, and the ring finger (RING) and RAWUL domains in RING1B and BMI1. Dashed boxes indicate protein regions built into the models. b, Gel filtration profile of reconstituted vPRC1 used for cryo-EM. c, Coomassie-stained SDS–PAGE showing vPRC1:Nuc and vPRC1:NucH2Aub1 samples used for cryo-EM. d, Cryo-EM reconstruction at 2.9 Å and final model of vPRC1:Nuc showing the RING:BMI1 ring finger heterodimer (gold and orange) bound to each face of the nucleosome disc (gray, DNA; pink, histone octamers). View orientation of the nucleosome relative to its dyad axis is indicated. e, Interaction between RING1B and the H2A acidic patch; the interacting residues are represented as sticks and contacts are marked by dashed lines. f, Cryo-EM reconstruction at 3.18 Å and model of vPRC1:NucH2Aub1 showing RYBP23–58 (blue) and ubiquitin (green) bound to each face of the nucleosome disc. View orientation of the nucleosome relative to its dyad axis is indicated. g, Focused refined map showing density of the RYBP zinc finger contacting ubiquitin and RYBP loop binding the nucleosome acidic patch. h, Interactions of RYBP23–58 with the H2A acidic patch and ubiquitin. Contacts between sidechains with clear density are represented as stick models and black dashed lines. Interactions with weak or unresolved density are indicated in gray and sidechains in pale colors. Acidic patch residues E60, E63, D70, N88, D89 and E91 are contacted by RYBP, whereas in e the same residues are contacted by RING1B.