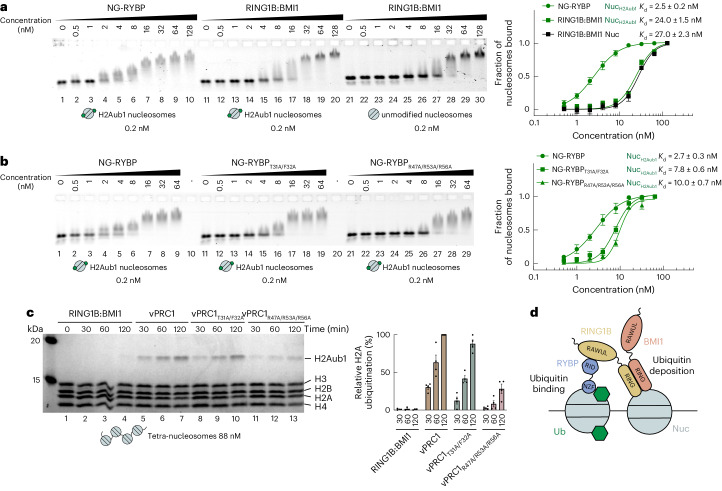
Fig. 2. High-affinity binding of RYBP to H2Aub1 nucleosomes via ubiquitin- and acidic patch contacts enables vPRC1 to generate H2Aub1 chromatin domains.
a, RYBP shows higher binding affinity than RING1B:BMI1 on H2Aub1 nucleosomes. Left, EMSA with the indicated concentrations of NG–RYBP or RING1B:BMI1 and 0.2 nM 647N-ATTO-labeled mononucleosomes that were H2Aub1-modified or unmodified as indicated. Right, quantitative analysis of EMSA data by densitometry of 647N-ATTO signal from independent experiments (n = 5), error bars (s.e.m.); apparent dissociation constant (Kd) values are indicated. b, Mutation of ubiquitin-contacting or acidic patch-contacting residues in RYBP reduces binding to H2Aub1 nucleosomes. Left, EMSA as in a, comparing binding of NG–RYBP, NG–RYBPT31A/F32A and NG–RYBPR47A/R53A/R56A with H2Aub1-modified mononucleosomes. Right, quantitative analysis from independent experiments (n = 3) as in a. c, Efficient H2A monoubiquitination by vPRC1 in nucleosome arrays relies on RYBP interaction with the nucleosome acidic patch. Left, ubiquitination reactions monitoring H2Aub1 formation by full-length RING1B:BMI1, wild-type vPRC1, vPRC1T31A/F32A or vPRC1R47A/R53A/R56A on tetranucleosomes after indicated incubation times, analyzed on Coomassie-stained 16% polyacrylamide gel. Right, quantification of H2Aub1 signal by densitometry from independent experiments (n = 4). In each experiment, the H2Aub1 signal in lane 7 was defined as 100% and used for quantification of H2Aub1 signals in other lanes on the same gel. Circles show individual datapoints with error bars (s.e.m.). d, Model of the vPRC1 read–write mechanism. Abbreviations as in Fig. 1a.