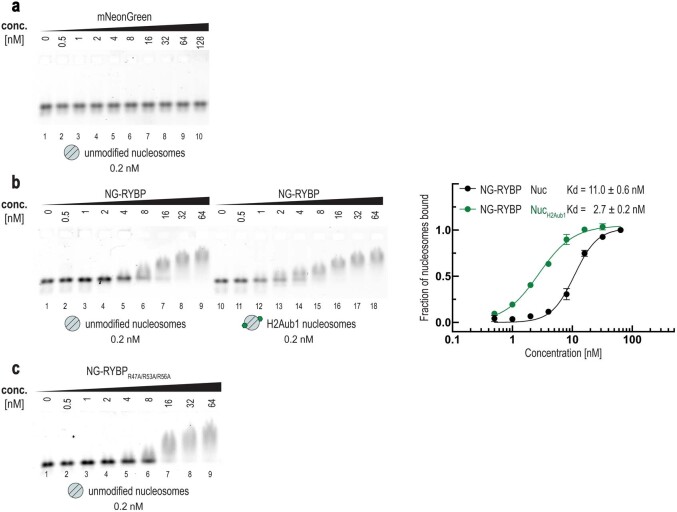
Extended Data Fig. 4. Binding of mNeonGreen, NG–RYBP or NG-RYBPR47A/R53A/R56A proteins to unmodified or H2Aub1-modified mononucleosomes.
(a) mNeonGreen (NG) protein lacks nucleosome-binding activity. Binding reactions with the indicated concentrations of NG and 0.2 nM 647N-ATTO-labeled unmodified mononucleosomes, analyzed by EMSA on 1.0% agarose gels. (b) NG–RYBP shows higher binding affinity for H2Aub1-modified nucleosomes than for unmodified nucleosomes. Left, binding assays by EMSA as in (a). Binding of NG–RYBP to H2Aub1-modified mononucleosomes as in Fig. 2a,b, is compared to the binding of the same wild-type NG–RYBP protein to unmodified mononucleosomes. Right, quantitative analysis of EMSA data by densitometry of 647N-ATTO signal from independent experiments (n = 3); error bars, SEM. The residual binding by the NG–RYBP protein on unmodified mononucleosomes in lanes 7–9 appears to be independent of RYBP-H2A acidic patch interactions because at high concentrations binding is also observed with NG–RYBPR47A/R53A/R56A on unmodified mononucleosomes (c); the nature of this RYBP interaction with nucleosomes is currently unknown.