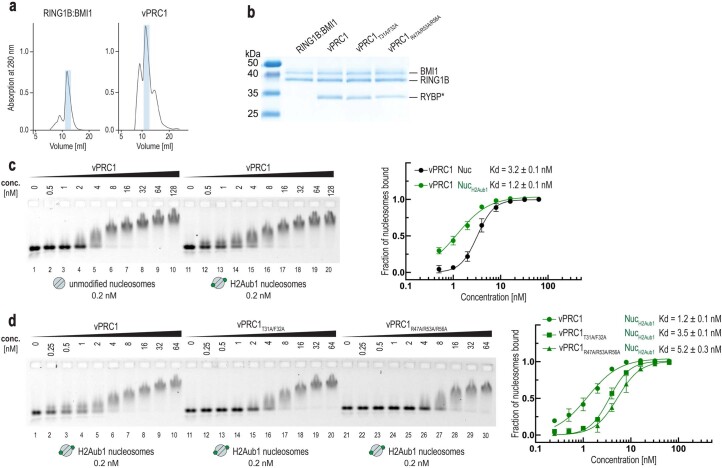
Extended Data Fig. 5. High-affinity binding of vPRC1 to H2Aub1-modified nucleosomes relies on RYBP–H2Aub1 contacts.
(a) Size-exclusion chromatography profiles of reconstituted RING1B:BMI1 full-length and vPRC1 wild-type complex (S200 column, absorption at 280 nm). Fractions used in all binding and activity assays are marked in blue. Note that the vPRC1 sample utilized for Cryo-EM analysis underwent an additional SEC run to assess the integrity of the complex (see Fig. 1b). (b) Coomassie-stained SDS gel showing the RING1B:BMI1 full-length dimer, wild-type and mutant vPRC1 complex samples used in biochemical assays. RYBP* denotes the wild-type or mutant form of RYBP, as indicated in the complex name above each lane. (c) vPRC1 shows higher binding affinity for H2Aub1-modified nucleosomes than for unmodified nucleosomes. Left, binding reactions with the indicated concentrations of vPRC1 and 0.2 nM 647N-ATTO-labeled unmodified or H2Aub1-modified mononucleosomes, analyzed by EMSA on 1.0% agarose gels. Right, quantitative analysis of EMSA data by densitometry of 647N-ATTO signal from independent experiments (n = 3); error bars, SEM. (d) RYBP interactions with ubiquitin and the H2A acidic patch account for the high binding affinity of vPRC1 complex on H2Aub1-modified nucleosomes. Left, binding assays by EMSA as in (c). Binding of wild-type vPRC1, vPRC1T31A/F32A and vPRC1R47A/R53A/R56A to H2Aub1-modified mononucleosomes. Right, quantitative analysis of EMSA results as in (c) from independent experiments (n = 3); error bars, SEM. Note that the binding observed at higher concentrations of wild-type or mutant vPRC1 (lanes 8–10, 18–20 and 28–30) likely represents the sum of binding interactions due to RING1B:BIMI1 Ring-finger-acidic patch contacts (Fig. 2a) and RYBP-nucleosome contacts unrelated to the RYBP–H2Aub1 interaction characterized in this study (Extended Data Fig. 4c).