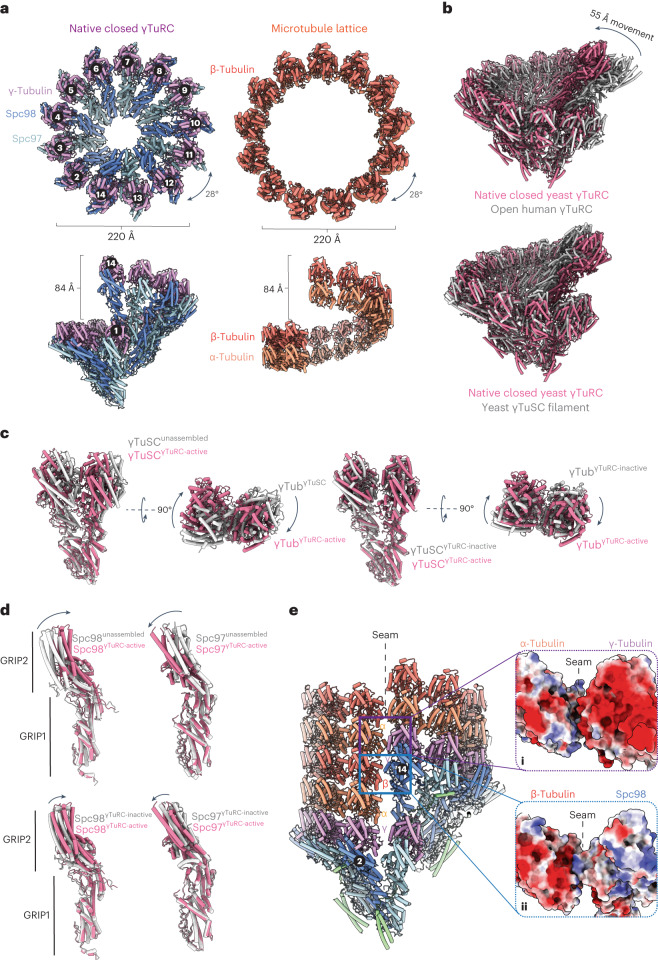
Fig. 2. γTuRC forms a perfect template for MT nucleation.
a, Top-down and side views of the native yeast γTuRC core complex (left) and the MT lattice (right) showing an identical helical geometry. The MT lattice was modeled on the basis of our 6.6 Å reconstruction of the native yeast MT lattice (Extended Data Fig. 1a). ‘(b)’ refers to the view shown in b. b, The native, closed and active yeast γTuRC structure was superimposed onto the inactive human γTuRC complex (PDB ID: 6V6S) (top) and single helical turn of the yeast γTuSC filament (PDB ID: 7m2x) (bottom), showing that γTuRC undergoes extensive conformational changes to match MT geometry and thus template MT nucleation. c, Left: a comparison of γTuSC geometries between unassembled γTuSC modules (PDB ID: 7m2z) and γTuSC modules in the context of active γTuRC. Right: a comparison of γTuSC geometries between γTuSC within the open-inactive γTuSC filament (PDB ID: 7m2x) and γTuSC in the context of active γTuRC. γ-tubulin pairs of each γTuSC module straighten up to adopt 13-PF MT geometry. Coordinates were superimposed on the Spc98GRIP1 domain. d, γ-Tubulin pairs straightening is accommodated by Spc98GRIP2 straightening and Spc97GRIP2 bending. PDBs were superimposed on the Spc98GRIP1 domain. e, A view of the interfaces at the γTuRC:MT seam, focusing on the electrostatic α-tubulin:γ-tubulin as well as Spc98:β-tubulin interfaces. The insets on the right show the α-tubulin:γ-tubulin (i) and Spc98:β-tubulin (ii) interfaces as electrostatic surfaces.