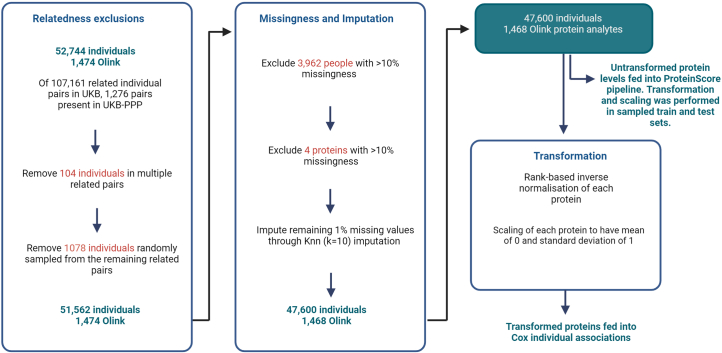
Extended Data Fig. 2. Summary of processing steps applied to the protein measurement data in UKB-PPP.
Related individuals were excluded, leaving a dataset containing 51,562 individuals with 1,474 Olink protein analytes measured. Next, 3,962 individuals that had >10% missing data were excluded, followed by four proteins that had >10% missing data. The remaining missing protein measurements (1% of total measurements) were imputed through K-nearest neighbours (Knn; k=10) imputation. The final dataset was comprised of 47,600 individuals and 1,468 Olink protein analytes. Protein levels were rank-based inverse normalised and scaled to have a mean of 0 and standard deviation of 1 prior to individual Cox PH analyses. Untransformed protein levels were fed into the model pipeline for ProteinScore development and were rank-based inverse normalised and scaled to have a mean of 0 and standard deviation of 1 in train and test sets separately once these were sampled for each outcome. Created with BioRender.com.