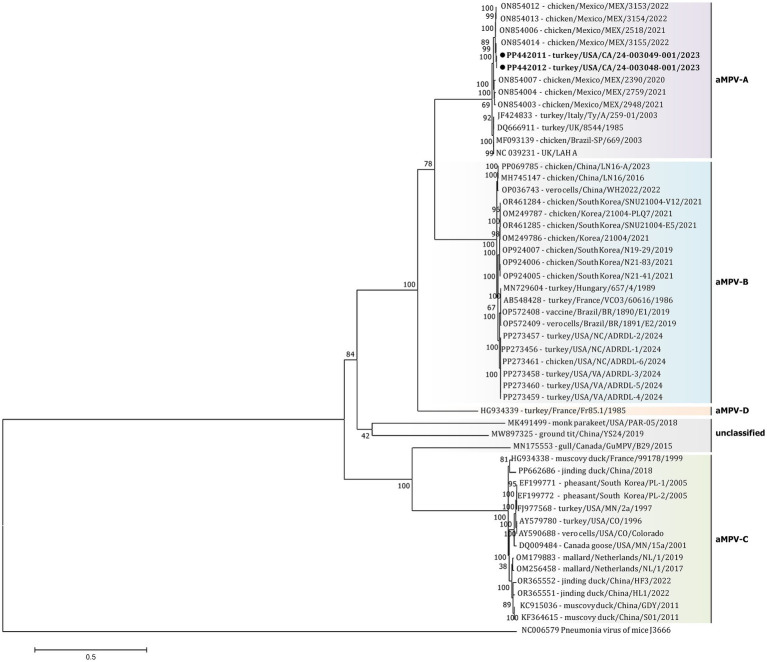
Figure 3.
Phylogenetic analysis of US aMPV-A strains 24–003048-001 and 24–003049-001 (marked with a black circle) within the genus Metapneumovirus based on complete nucleotide genome sequences. Strain names include GenBank accession numbers, along with host species, country origin, strain name, and year of sample collection. The tree was constructed using the Maximimum-Likelihood method based on the best-fitting General-Time Reversible (GTR + G + I) substitution model with 1,000 bootstraps. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 52 nucleotide sequences (sequence from Murine orthopneumovirus (NC_006579) was included as an outgroup). All positions containing gaps and missing data were eliminated. There was a total of 10,787 positions in the final data set.