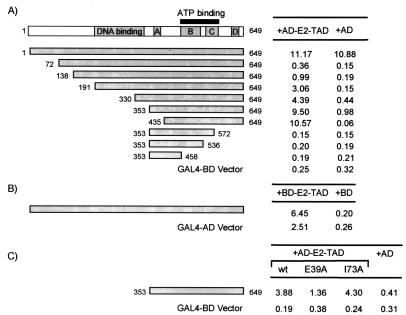
FIG. 1.
Interaction of E1 with the TAD of E2 in the yeast two-hybrid system. (A) Diagram of the E1 protein (top). The shaded boxes labeled A, B, C, and D represent regions of E1 that have high sequence similarity with large T antigens of SV40 and polyomaviruses (11). A region that is essential for DNA binding (see the introduction) is also shown as a shaded box. The solid box indicates the position of the ATP-binding domain of E1. β-Galactosidase activity detected in yeast cells expressing either E1 or a fragment of E1 fused to the GAL4 DNA-binding domain (BD) or the GAL4 BD alone and also expressing either the E2 TAD (amino acids 1 to 209 of E2) fused to the GAL4 activation domain (AD) or the GAL4 AD alone is shown below. The portions of E1 fused to the GAL4-BD are diagrammed as shaded boxes. The amino acids at the N and C terminus of each E1 fragment are indicated. (B) β-Galactosidase activity detected in yeast cells expressing either the E2 TAD fused to the GAL4 BD or the GAL4 BD alone and also expressing either E1 fused to the GAL4 AD or the GAL4 AD alone. (C) Effect of substitutions in the TAD of E2 on interaction with E1. β-Galactosidase activity detected in yeast cells expressing either E1 (amino acids 353 to 649) fused to the GAL4 BD or the GAL4 BD only and also expressing either the wild-type (WT) E2 TAD or a mutant E2 TAD (E39A or I73A) fused to the GAL4 AD or the GAL4 AD alone. The numbers on the right indicate the amounts of β-galactosidase activity (in Miller units) measured in S. cerevisiae for each combination of plasmids.