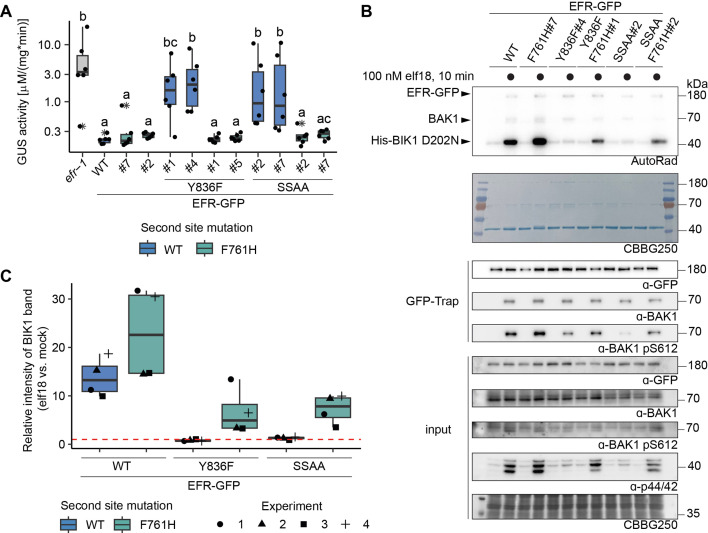
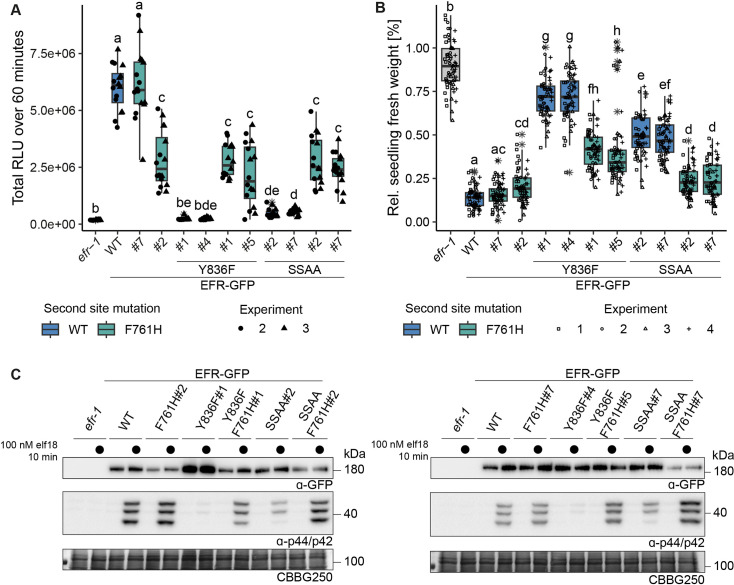
Figure 3. EFRF761H/Y836F and EFRF761H/SSAA recover receptor complex activation.
(A) In infection assays, GUS activity was high in the positive control efr-1 line. GUS activity level was reduced in the EFRWT and EFRF761H complementation lines, but much less so in the EFRY836F and EFRSSAA complementation lines. By contrast, EFRF761H/Y836F and EFRF761H/SSAA complementation lines displayed substantially repressed GUS activity. Each experiment was repeated three times with similar results. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (P=5.704*10–7), Dunn’s post-hoc test with Benjamin-Hochberg correction (P ≤ 0.05) Groups with like lowercase letter designations are not statistically different. (B) In IP kinase assays, ligand-induced interaction of EFRWT and EFRF761H with BAK1 increased transphosphorylation of BIK1D202N, but this was abolished for EFRY836F and EFRSSAA. Both EFRF761H/Y836F and EFRF761H/SSAA showed partially restored BIK1D202N trans-phosphorylation as well as BAK1 S612 phosphorylation (across four replicates for EFRF761H/SSAA and in two out of four replicates for EFRF761H/Y836F). Samples were also probed for MAPK phosphorylation for effective ligand treatment. Treatment: 100 nM elf18 for 10 min. (C) Quantification of BIK1D202N band intensity observed in autoradiographs from the four independent replicates performed. Dotted red line indicates unchanged band intensity in mock vs. elf18 treatment.