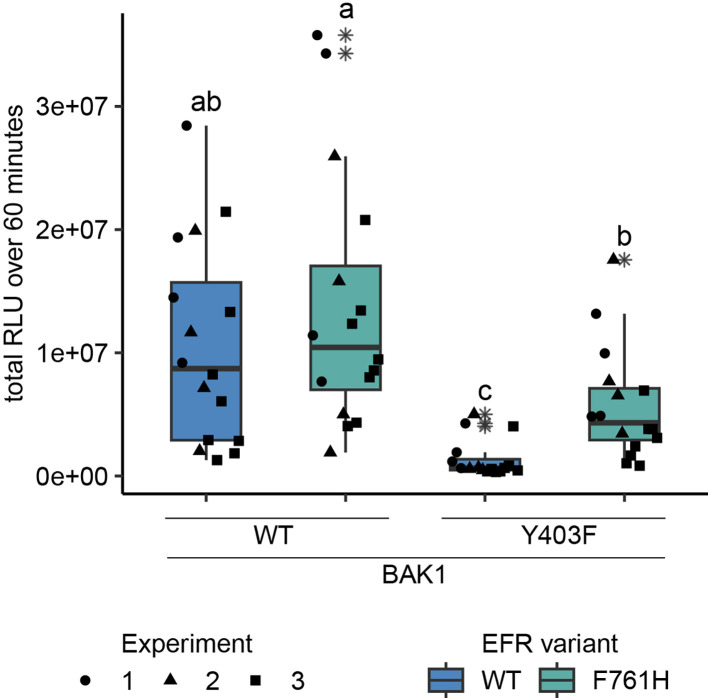
Figure 4. EFRF761H recovers BAK1Y403F function.
The cytoplasmic domains of BAK1 and EFR variants with fused RiD-tags were transiently expressed in N. benthamiana and leaf discs were treated with Rap to induce dimerization. EFR and EFRF761H induced a similar total oxidative burst when BAK1 was co-expressed. The co-expression of BAK1Y403F and EFR diminished the oxidative burst, which was restored partially when EFRF761H was co-expressed. Outliers are indicated by an additional asterisk and included in statistical analysis. Statistical test: Kruskal-Wallis test (p<8.516 *10–7), Dunn’s post-hoc test with Benjamin-Hochberg correction (p ≤ 0.05) Groups with like letter designations are not statistically different.