Figure 1.
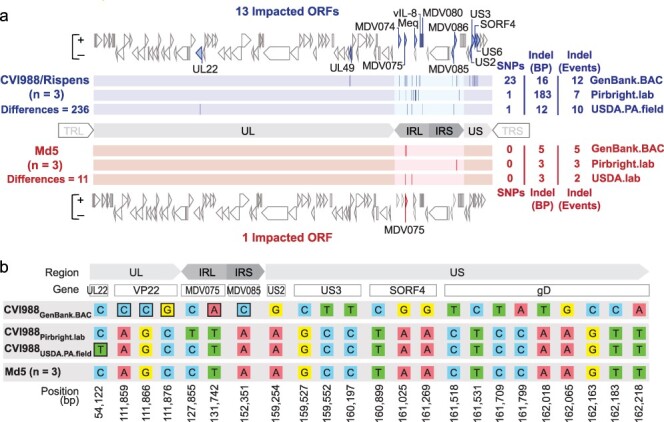
Comparative analysis of CVI988 and Md5 consensus genomes. (a) A total of three trimmed consensus genomes (TRL and TRS removed) were obtained for each strain (CVI988 = Blue, Md5 = Red) and aligned using MAFFT. Consensus-level differences including SNPs and Indels are highlighted (dark lines) based on their relative locations in each respective genome. The total number of differences between genomes of the same strain is indicated on the left, with a breakdown for each individual genome indicated on the right; these loci are all listed in detail in Supplementary Tables S2 and S3. For each strain, ORFs with consensus-level differences in at least one of the three genomes belonging to that strain are highlighted and labeled (with alternative names when possible). The total number of ORFs with consensus-level differences is 13 for CVI988 and 1 for Md5. (b) ORF-associated SNPs were found between consensus genomes of CVI988. Genomic positions are indicated relative to CVI988GenBank.BAC, which accounted for 21 out of 23 ORF-associated SNPs. Completely unique SNPs that were not found in any of the other 38 MDV genomes included in the study are highlighted (black borders).
