Figure 4.
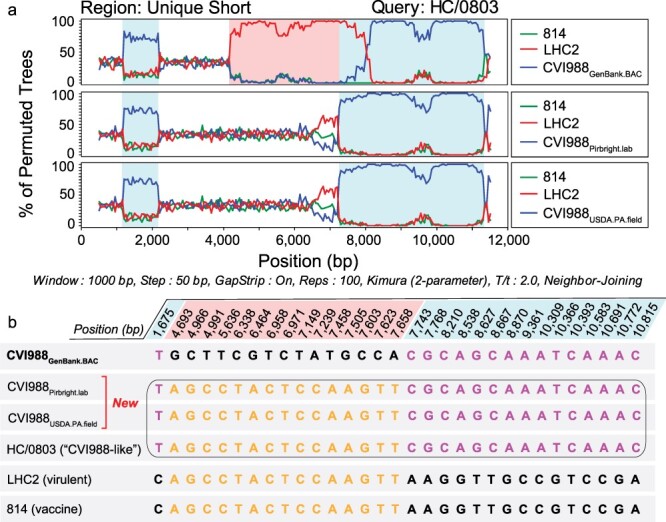
Recombination analyses of MDV strains previously described as natural recombinants of CVI988. (a) Bootscan analyses of the unique short region of “CVI988-like” MDV strain HC/0803 using SimPlot. Additional strains include 814 (vaccine strain), LHC2 (virulent strain; both from China), CVI988GenBank.BAC (top), CVI988Pirbright.lab (middle), and CVI988USDA.PA.field (bottom). Nucleotide positions are indicated relative to the US region of CVI988GenBank.BAC. (b) The FindSites tool in SimPlot was used to identify permuted trees and informative sites associated with recombination events in the US region of HC/0803. When CVI988GenBank.BAC was used for the analysis, two permuted trees were detected. Each permuted tree was associated with 15 informative sites (shaded light blue or light red, depending on their respective permuted tree). By contrast, both CVI988Pirbright.lab and CVI988USDA.PA.field shared 100% identity with “CVI988-like” variants across all 30 positions (black border). This resulted in only a single permuted tree and no recombination events being detected. See Supplementary Fig. S1 for related analyses of additional strains.
