Figure 5.
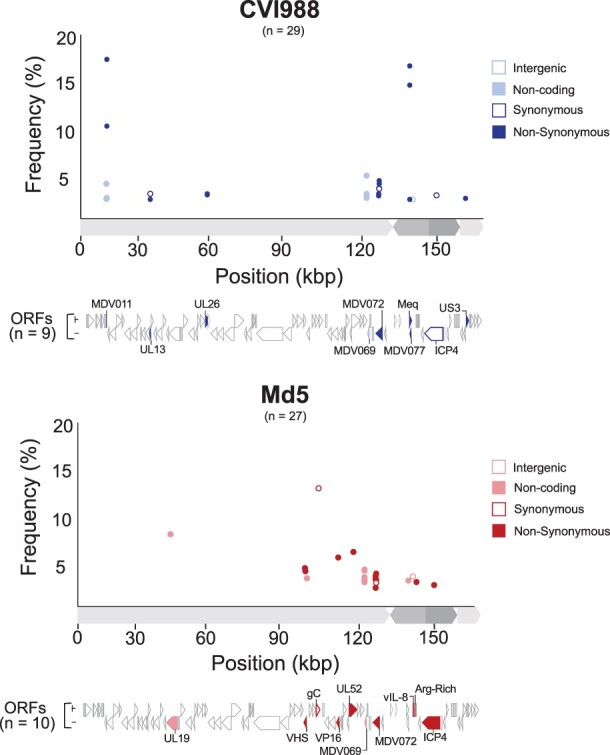
Minor variant distribution in Pirbright CVI988 and Md5 populations. For each strain (CVI988 = blue/top panel, Md5 = red/bottom panel), a single viral population was ultra-deep sequenced (>1000×) to enable identification of minor variants (Table 1). Positions outside of tandem repeats and homopolymers where the minor allele was present in ≥2% of the reads were identified as minor variants. Each minor variant was classified depending on its relative location to ORFs and its impact on the resulting amino-acid sequence (when applicable). ORFs associated with minor variants are labeled and highlighted beneath the graph using the same criteria.
