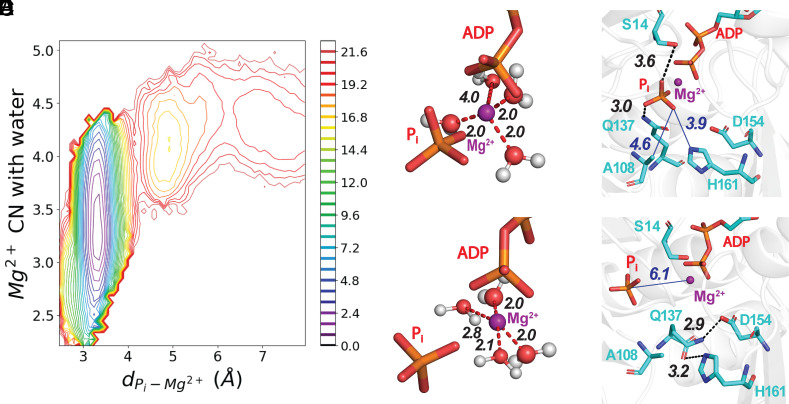
Fig. 2.
The release of Pi proceeds with the solvation of Mg2+ and the disruption of interactions with nearby residues in the active site. (Panel A) A 2-dimensional plot of PMF as a function of the number of waters coordinated with Mg2+ and the distance between Pi and Mg2+ calculated from one release trajectory. The bar on the Right has the color code for energy levels in kcal/mole. SI Appendix, Fig. S4 has PMFs from five other trajectories. (B–E) Stick figures of the active site with Mg2 depicted as a purple sphere using the VDW presentation with a 0.8 scale, hydrogen bonds as black dashed lines, electrostatic interactions as red dashed lines, and nonbonding distances as blue solid lines. (B and C) ADP and Pi at two time points during a WT-MetaD simulation. (Panel B) Mg2+ is coordinated with three water molecules. (Panel C) Mg2+ coordinated with four water molecules after losing coordination with Pi. (D and E) Hydrogen bonds at two time points during a WT-MetaD simulation. (Panel D) Before dissociating from Mg2+, Pi was coordinated with the oxygen of the side chain of residue S14 and a hydrogen atom of the side chain amine group of residue Q137. (Panel E) After changing its rotameric conformation, the side chain of Q137 forms a hydrogen bond between its side chain oxygen and a hydrogen atom on the ring of H161. During our MD simulations, the reorientation of Q137 always occurs before Pi loses its coordination with Mg2+ (see SI Appendix for more details).