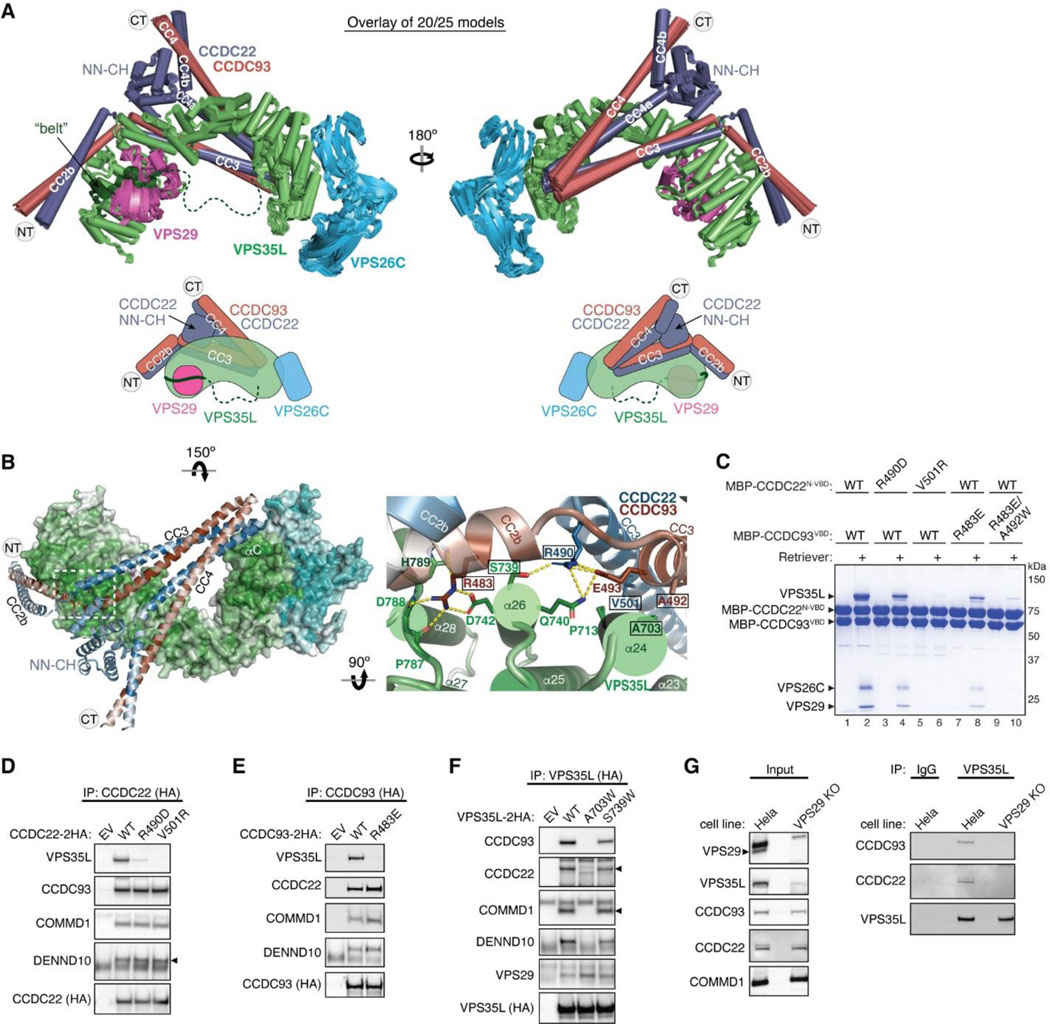
Fig. 5. Structural model of CCDC22-CCDC93 binding to Retriever.
(A) Overlay of AlphaFold Multimer models and schematic showing Retriever binding to CCDC22-CCDC93. For clarity, inconsistent models (5 out of 25 total models) are excluded. Unreliable structural regions showing inconsistency between models and high PAE scores are removed, including the peptide linker following the “belt” sequence in VPS35L (dotted green line). (B) Interaction surface between Retriever and CCDC22-CCDC93 colored by conservation score using the same scheme shown in Fig. 2. Key interactions are shown as sticks and polar interactions are represented with a dashed yellow line. Residues mutated in this study are outlined with a black box. (C) Coomassie blue-stained SDS PAGE gel showing indicated variants of MBP-CCDC22 NN-CH-VBD/MBP-CCDC93 VBD dimers (200 pmol) pulling down Retriever (60 pmol). (D-F) Immunoprecipitation of indicated mutants of CCDC22 (D), CCDC93 (E), and VPS35L (F) expressed in HEK293T cells and immunoblotting of indicated proteins. (G) Immunoprecipitation and immunoblotting of VPS35L from parental HeLa cells and a VPS29 knockout line derived from these cells.