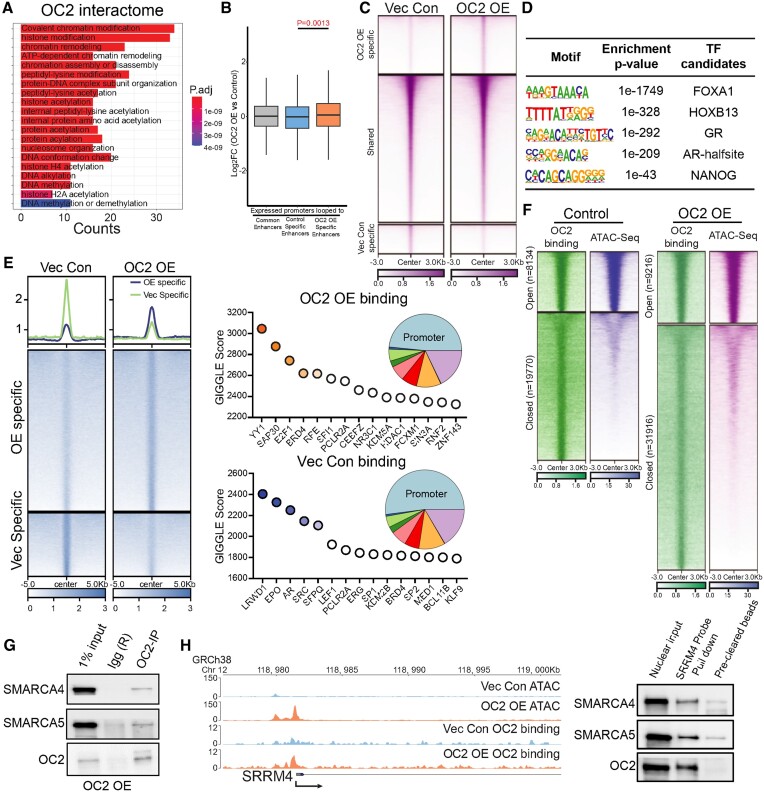
Figure 2.
OC2 interacts with CRCs and alters chromatin accessibility. (A) Gene Ontology analysis of the OC2 interactome proteins identified by IP-MS experiments (N = 2) showed enrichment of chromatin remodeling complexes. (B) HiC-seq was integrated with histone H3K27Ac CUT&RUN-seq to identify specific enhancers in LNCaP Vec Con and OC2 OE cells (N = 2). RNA-seq was then integrated to access the specific enhancer looped those upregulated gene promoter regions, suggesting the newly formed looping structures contributed to OC2-directed gene expression changes. (C) Normalized tag densities for ATAC-seq in LNCaP Vec Con and OC2 OE cells showed OC2 induced chromatin remodeling (N = 2). (D) Motif enrichment analysis for hyper-accessible regions in OC2 OE from ATAC-seq data. (E) Normalized tag densities for the OC2 cistrome in LNCaP Vec Con and OC2 OE cells through CUT&RUN-seq (Left). GIGGLE analysis for TF binding similarity in the OC2-specific cistrome in both conditions (Right) (N = 2). (F) Normalized tag densities for integrated analysis of ATAC-seq and OC2 CUT&RUN-seq. 75% of the OC2-bound regions consisted of closed chromatin. (G) IP-WB showed OC2 interacts with SMARCA4 and SMARCA5. (H) ATAC-seq suggested OC2 binds to the chromatin and induced chromatin opening in the SRRM4 promoter region (left). DNA affinity precipitation assay (DAPA) showed binding of SMARCA4 and SMARCA5 to the SRRM4 promoter region under OC2-enforced conditions (right) (Refer to methods).