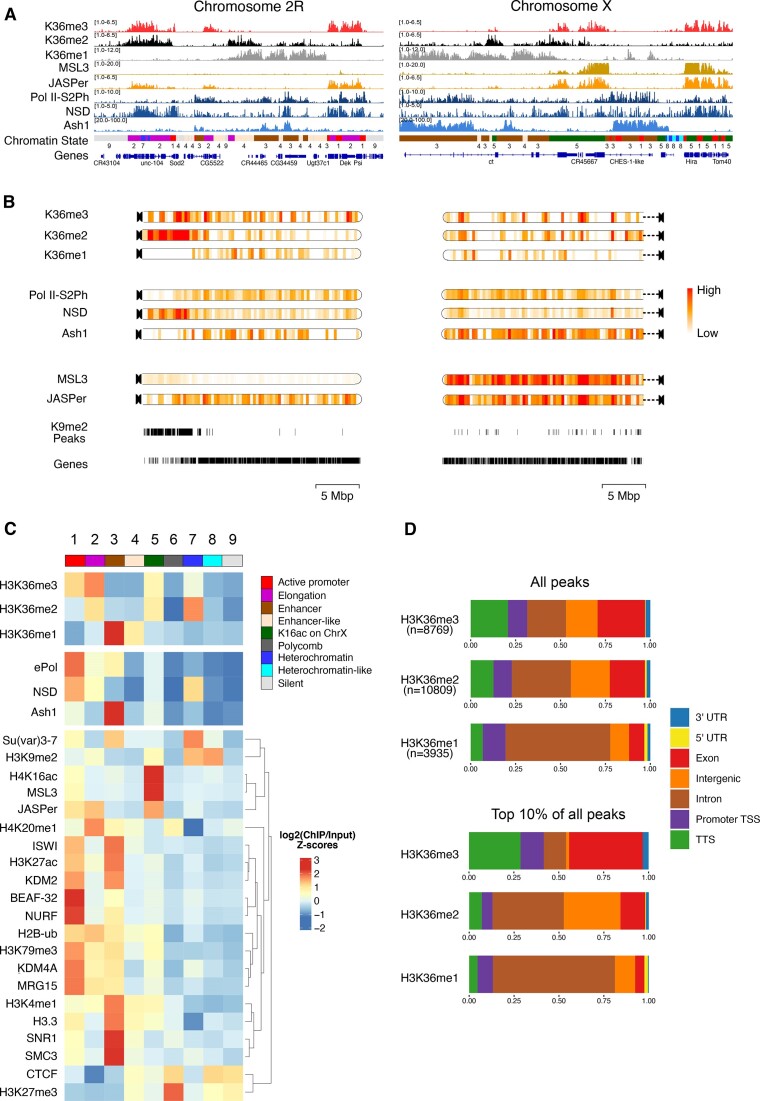
Figure 1.
H3K36 modifications and HMTs mark distinct chromatin states in male Drosophila cells. (A) Genome browser profiles for representative Drosophila chromosomes 2R and X of spike-normalized MNase-ChIP of K36me1/2/3, the K36me3 reader proteins JASPer and MSL3 and the HMTs NSD and Ash1. ‘ePol’ refers to the signal generated by an antibody against RBP1-S2ph, as a proxy for RNA-polymerase-S2ph-interacting Set2. The Ash1 profile was taken from (41). The 9-state ChromHMM (modENCODE) is color-coded and explained in (C). (B) ChromoMaps representing steady-state enrichment of K36me1/2/3, HMTs and K36me3 readers in 10-kbp genomic bins for chromosomes 2R and X. Scale bars are different for each chromoMap to facilitate visualization of changes. The individual scale values for each state and chromosome are shown in Supplementary Figure S1. Tracks representing H3K9me2 peaks and annotated genes serve as a reference for mappable pericentric heterochromatin domains and transcribed chromatin, respectively. (C) Chromatin State enrichment (9-state ChromHMM, (2)) for K36me1/2/3, HMTs and K36me3 readers. Published ChIP-seq/CUT&RUN profiles of histone modifications and other chromatin proteins were hierarchically clustered to highlight differences between chromatin states. (D) Genomic features marked by either all K36me1/2/3 domains (top) or filtered for the strongest 10% (bottom).